Summary
Many of our organs can maintain and repair themselves during homeostasis and injury, due to the action of tissue-specific, multipotent stem cells. However, recent evidence from mammalian systems suggests that injury stimulates dramatic plasticity, or transient changes in cell potential, in both stem cells and more differentiated cells. Planarian flatworms possess abundant stem cells, making them an exceptional model for understanding the cellular behavior underlying homeostasis and regeneration. Recent discoveries of cell lineages and regeneration-specific events provide an initial framework for unraveling the complex cellular contributions to regeneration. In this review we discuss the concept of cellular plasticity in the context of planarian regeneration, and consider the possibility that pluripotency may be a transient, probabilistic state exhibited by stem cells.
Keywords: Stem cells, regeneration, homeostasis, planaria, plasticity
Cell dynamics during regeneration
Stem cells are cells that divide continuously, both to self-renew and to produce various cell types in our bodies. During embryonic development, stem cells are multipotent, capable of producing all the cell types in the animal. As development proceeds, stem cell potential gradually diminishes, eventually becoming lineage-restricted and producing only a subset of cell types matched to the organ [1]. Present throughout our lives, these tissue-specific stem cells replenish dying cells and maintain the physiological function of our organs, in a process called homeostasis.
Based on their low-level activity in adult animals, tissue-resident stem cells participate in tissue repair by producing the particular cell types present in a given organ. However, emerging evidence suggests that while these stem cells can occasionally participate in tissue repair, the changing environment induced by wounding can also stimulate other, differentiated cells to contribute to regeneration [2,3]. This expansion of lineage potential – termed plasticity – has been described in several mammalian organs, including mammary glands, prostate glands, lung, the small intestine, and hair follicles [4–9]. For example, in the murine small intestine, homeostasis is largely driven by fast-cycling cells located at the crypt base [10]. Wounding or genetic ablation of these rapidly dividing cells causes the typically unipotent quiescent stem cells at the +4 position to become multipotent, now producing all of the cell types comprising the crypt [11]. Similarly, lineage-tracing of cells expressing the differentiated marker Dll-1 normally produce Paneth, enteroendocrine and secretory goblet cells, but after ablation of stem cells following irradiation, they now give rise to long-lived, multi-lineage clones [12]. Severe injuries in the lung also cause differentiated cells to adopt proliferative behavior and restore damaged tissues [13,14]. Therefore, injury induces environmental stimuli that elicit distinct cellular behaviors, facilitating organ repair.
In general, the dynamic behavior stimulated by injury calls into question our formal definitions of “stem cells” and “differentiated cells”, and suggests that the differentiated state, at least for certain tissues, may not be terminal. Instead, within mature organs, cells may adopt what could be considered a stable differentiation state, permitting plasticity to occur upon injury. Recent advances in the transcriptional analyses of single cells underscore this possibility, as these data have now revealed a striking degree of single-cell, non-genetic heterogeneity in otherwise apparently homogeneous cell populations [15–19]. Such heterogeneity likely reflects changes in the dynamics of expression of key regulatory genes. In light of this growing evidence, the traditional definition of “cell type” may be in need of reevaluation.
The flatworm Schmidtea mediterranea as a system for studying in vivo stem cell behavior
Freshwater planarians are an exceptional model organism for studying the in vivo regulation of stem cells and how they contribute to regeneration [20]. Planarians can regenerate virtually any body part after amputation, due to the involvement of pluripotent stem cells (neoblasts) that are dispersed throughout the body. In this review we argue that the greater regenerative capacity of planarians offers a tremendous opportunity to understand the cellular mechanisms underlying regeneration, including the interplay between differentiated tissues and stem cells, and transitions between homeostatic and regenerative states.
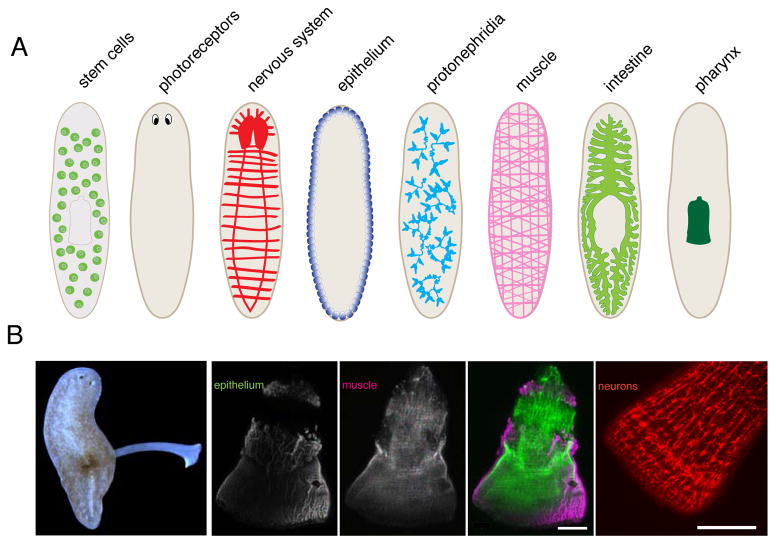
Despite their relatively simple outward appearance, planarian anatomy is quite elaborate (Figure 1), consisting of derivatives of all three germ layers. Planarian organ systems include a complex central nervous system [21,22], photoreceptors [23], a digestive system consisting of a branched gastrovascular system [24], a pharynx, and a primitive excretory system called protonephridia [25,26], all enveloped by body wall muscle and epithelial cells. All of these organs regenerate readily after amputation.
Figure 1.
Planarian anatomy. (A) Various organs in asexual flatworms. Each organ illustrated here consists of several cell types. (B) Left, live animal extending its pharynx. Right, pharynx anatomy in isolated pharynges with stained epithelial cells, muscle, neurons (α-FMRF-amide). Scale bars, 100μm.
Distributed throughout the animals are small, dividing cells called neoblasts. Thought to be the only dividing cells in the animal, neoblasts produce various cell types based on lineage tracing [24,27–29] and uniformly express many markers including the Argonaute protein piwi-1 and histone H2B [30,31]. Aspects of the molecular regulation of stem cells have been extensively reviewed elsewhere [32,33]. Recently, transplantation assays were developed to determine the differentiation potential of single stem cells in irradiated hosts (which lack stem cells). With time, single cells produced all of the animal’s tissues, formally demonstrating that one neoblast (termed “cNeoblast”, for clonogenic Neoblast) can be truly pluripotent [29]. However, rescue occurs at a low frequency (7/120 transplanted cells), and it is unclear whether this reflects a relatively low, natural occurrence of cNeoblasts, or is a result of technical limitations. Furthermore, molecular markers for these cells have not yet been identified, leaving questions about their physical location and behavior unresolved.
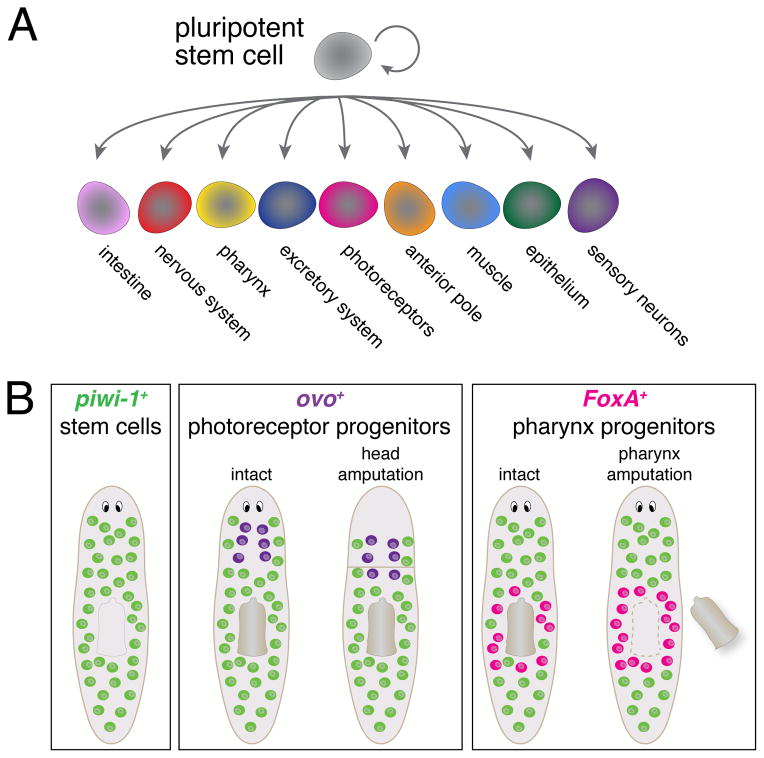
The abundance of stem cells, their broad distribution, and their ability to produce so many different tissue types suggests that this is a heterogeneous cell population. Measurements of gene expression in isolated stem cells [34–36] and in vivo [37] have confirmed the heterogeneity of neoblasts, and some heterogeneously expressed genes are transcription factors essential for organ regeneration. These have been identified by transcriptional profiling of mature tissues [38,39], testing candidates [23,28], RNAi screening for organ regeneration defects [25,40], gene expression analysis of neoblasts [36] or RNA-sequencing purified neoblasts [41] (Figure 2A). Knockdown of these organ-specific transcription factors causes a failure to regenerate the cognate organ, indicating that these genes may function to mark the future fate of stem cells. Coexpression of these lineage markers in subsets of neoblasts (broadly defined as piwi-1+) raises the possibility that lineage specification occurs at the level of the stem cell. Therefore, the stem cell population may be fragmented into lineage-restricted subsets [42], but the significance of heterogeneity, and how it may contribute to regeneration, is not yet clear.
Figure 2.
Lineage progenitors for organ regeneration. (A) Several specialized progenitors for many tissues are produced during homeostasis, derived from pluripotent stem cells. (B) Changing distribution of organ-specific progenitor markers after amputations (highlighted in pink or purple).
Stem cell lineages during homeostasis
During homeostasis, neoblasts undergo steady rates of mitosis, replacing cells lost to the normal wear and tear of adult life. Unlike other invertebrate model systems, planarians can grow (or shrink) to any size depending on feeding status [20,43,44]. Growth is fueled by stimulating stem cells to proliferate [45], and is counterbalanced by a constant rate of cell death, maintaining body size [46]. As they experience cycles of growth and degrowth, overall scaling of organs is preserved [47–49], suggesting that all cell types are produced (or lost) in a proportional manner [50–54].
Lineage tracing experiments show that during homeostasis, stem cell progeny are steadily incorporated into existing organs including epithelial cells, intestine and neurons [24,27,29,55]. As these cells are produced, they stably express tissue-specific transcripts. Therefore, a general assumption is that during normal steady-state conditions, pluripotent cells produce various specialized progenitors at a constant rate, perhaps stochastically. Now that several progenitor markers are known, a quantitative analysis of the lineages generated by stem cells during homeostasis will resolve this issue. An intriguing possibility is that during homeostasis, the extensive diversity of transcription factors expressed within neoblasts may be explained by dynamic and possibly transient expression of determinants across the stem cell pool over time. It is not yet known, however, whether the presence of stable progenitor cells influences the behavior of individual stem cells during homeostasis.
Organ regeneration highlights dynamic stem cell responses
The presence of progenitor markers within the stem cells creates an opportunity to understand how regeneration alters the dynamics of these cell populations. Because most organs in planarians are broadly distributed throughout the body (Figure 1), amputation typically removes only part of the organ, leaving the remainder damaged. This incomplete organ loss confounds analysis of progenitor behavior. Two examples of progenitor markers for anatomically restricted organs offer an opportunity to observe how stem cells contribute to de novo organ regeneration.
Photoreceptors are located in the anterior of the animal, and the transcription factor ovo is required for their regeneration [38]. During homeostasis, these ovo+/piwi-1+ progenitors are produced in a steady stream and are expressed in vanishingly few neoblasts directly posterior to the photoreceptors. Head amputation stimulates the production of new photoreceptors, initiated by activation of ovo within stem cells (Figure 2B). Knockdown of ovo causes a highly specific inability to regenerate all cell types within the photoreceptors, while other tissues regenerate normally, suggesting that ovo functions as a selector gene [56] to drive photoreceptor formation.
The pharynx can be completely removed without overt damage to other tissues, facilitating the study of the stem cell response to selective organ loss. The Forkhead transcription factor FoxA is essential for regeneration of the pharynx but not other organs [40,41]. FoxA is expressed in a subset of stem cells, but only those in the immediate vicinity of the pharynx (Figure 2B). After chemical amputation, stem cells activate expression of FoxA specifically within stem cells. Although the mechanism of FoxA upregulation is not known, the increase in the proportion of stem cells expressing FoxA suggests that a dynamic shift in gene expression in progenitor cells is induced by organ loss.
Amputation of heads or tails (as in Figure 3) generates fragments lacking pharynges. In contexts where de novo regeneration of the pharynx occurs in fragments, stem cells in the anterior or posterior (not normally expressing FoxA) activate its expression [57] (and Adler, unpublished), indicating plasticity in the stem cells during regeneration. For both photoreceptors and the pharynx, pre-existing stem cells in the body must recognize the absence of these organs, and subsequently initiate expression of organ progenitors. This highlights key questions in planarian regeneration: are stem cells constantly monitoring the presence and absence of every organ? If so, how are the stem cells instructed to produce specific organs after amputation? Answering these questions are important future goals of planarian research.
Figure 3.
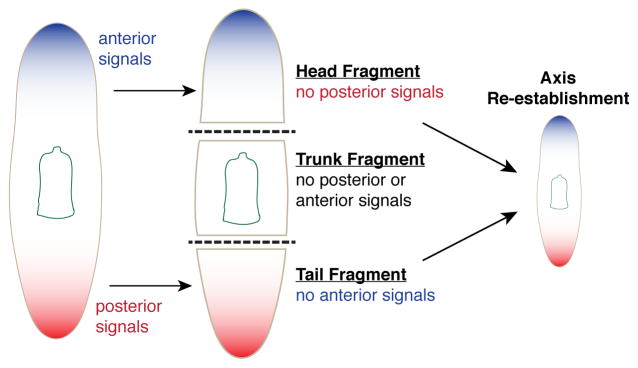
Amputation initiates polarity re-establishment. Removal of anterior and posterior regions induces dramatic changes in the animal. Transcriptional markers of the anterior and posterior (represented by blue and red, respectively) are expressed in differentiated cells, and removed after significant amputations, leaving disproportional axial patterning in fragments. Within 12–24 hours, expression of these markers is reestablished in fragments and instructs fate determination of regenerating tissues.
Injury-induced Responses in Planarians
After injuries, planarians rapidly heal wounds and quickly re-establish proper axial polarity. Tissues are regenerated within days, almost without error, after almost any type of amputation [24,58,59]. Organs, transcripts, and antigens that are normally restricted to specific regions now redistribute to establish proper proportioning of the animal [57,60,61]. Wounding induces potent changes throughout the animal, in both differentiated tissues and in stem cells, significantly altering the organismal context in which differentiated and stem cells reside [62,63]. Understanding how injury induces this transition from a homeostatic state into a regenerative state, and how modified stem cell behavior contributes to regeneration are key issues to resolve.
Immediate transcriptional changes in differentiated tissues
Although the identity of a wound signal is unknown, it likely diffuses quickly because within minutes of injury, dramatic transcriptional changes ensue throughout the animal [64,65], in both stem cells and differentiated tissues. Analysis of these early-response genes, mostly present in differentiated cells, showed distinct classes of wound-response genes. Probably because of functional redundancy, knockdown of many of these genes do not cause detectable phenotypes [64], leaving some uncertainty as to their function in regeneration. However, these early transcriptional changes in the pre-existing tissue are essential for initiating regeneration.
Many patterning genes are expressed in distinct domains during adult homeostasis. Removal of heads or tails disrupts this normal pattern, inducing disproportional expression patterns (Figure 3) [66]. After amputation, these markers reappear in a stereotypical manner at anterior-facing (sFRP-1, wnt2 and sFRP-2) or posterior-facing (wntP-2) wounds, even in the absence of stem cells [63,67,68], suggesting that these potential morphogens could provide axial information during regeneration. In fact, grafting of stem-cell-depleted tissue influences the fate of regenerated tissues [69,70]. Thorough analysis of several potential positional markers shows that they are expressed subepidermally, in subsets of muscle cells [71], and may create a coordinate system for regeneration, providing positional information by transcriptional activation of patterning genes [72]. Therefore, differentiated cells actively participate in regeneration by expressing patterning genes that directly influence cell fate decisions.
Stem cell response to wounding
Stem cells also respond rapidly to injuries. After wounding, two bursts of cell proliferation occur: six hours after injury, a wave of proliferation spreads throughout the entire body, and then two days after amputation, there is a localized peak of proliferation near the wound site [73–75]. Transplantation assays show that wounding mobilizes stem cell migration toward the amputation plane [76]. Transcriptional profiling with microarrays [64] and RNA-sequencing of purified stem cells [41] also identify several transcription factors that are upregulated in the first two days of regeneration.
Transcriptional plasticity of stem cells is best understood in the formation of a new anterior pole during regeneration. After removal of anterior tissues, neoblasts near the wound site initiate a cascade of several transcription factors, beginning with the Forkhead transcription factor FoxD [77,78] and then Zic-1, which further specifies subpopulations of stem cells to generate an anterior pole [79]. Anterior pole cells (which are differentiated cells) express several genes required for normal head formation: notum, follistatin, prep, and pbx [78–81]. To promote head formation, Wnt signaling must be suppressed, which is achieved by expression of the Wnt inhibitor notum. The rapid response of pre-existing tissue to activate this cascade of transcription factors demonstrates the plasticity of stem cells to wounding.
The signaling machinery responsible for altering stem cell behavior is poorly understood. Some potential receptors have been identified by transcriptional profiling or candidate approaches, such as the FGF receptor fgfr-1 [31], the EGF receptor efgr-3 [82] and the G-protein-coupled receptor P2X-A [83]. Future efforts will extend our knowledge of the signaling machinery regulating stem cell behavior, as well as whether they instruct specific responses (e.g. inducing proliferation or differentiation).
The end of regeneration and the return of homeostasis
Regeneration is expected to invoke distinct biological responses as compared to the homeostatic state. This is supported by evidence that knockdown of some genes impairs regeneration and not homeostasis. For example, the Bmp inhibitor follistatin [80,84] and the transcription factor runt-1 [64] exhibit regeneration-specific defects, while homeostasis is unaffected. Other genes, including the growth-regulatory kinase Tor and JNK, decouple rates of proliferation and apoptosis normally observed during regeneration [85,86].
Therefore, stem cells may adopt two distinct states – homeostasis and regeneration – and each state may exhibit different transcriptional and cellular characteristics. In adult animals, most of our organs reach steady state after development completes [87]. However, for organs that regenerate readily (such as blood, liver, and intestine), a feedback mechanism may exist to limit organ size and prevent excessive overgrowth [88]. Without this inhibition, these organs may undergo unlimited growth, potentially resulting in a cancerous state. This feedback control could be either intrinsically or extrinsically regulated [89], and the mechanism may differ among organs. Because planarians’ bodies continually expand and shrink during their lives, these types of organism-wide signals may exhibit heightened activity throughout a planarian’s lifetime. It will be interesting to determine which cell types may be involved in sensing tissue loss and alters its behavior after amputation.
One appealing hypothesis is that mature organs produce specific factors that inhibit their own growth [90]. The most well-characterized example of these types of signals is the TGFβ family member myostatin, which is expressed in mammalian muscle; mutations in myostatin cause vast muscle overgrowth [91,92]. In planarians, distinct, localized organs like the pharynx and photoreceptors could produce this kind of signal. In fact, some evidence exists that the pharynx can inhibit its own regeneration [93–95]. How might this signaling be regulated? For example, are stem cells constantly monitoring the presence of every differentiated cell type in the animal? Because cNeoblasts do not increase their proliferative rates after amputation [29], these cells are unlikely to sense the absence of organs. Instead, this responsibility may fall on the organ-specific progenitors embedded in the stem cell population.
Redefining regenerative potential as a cellular state
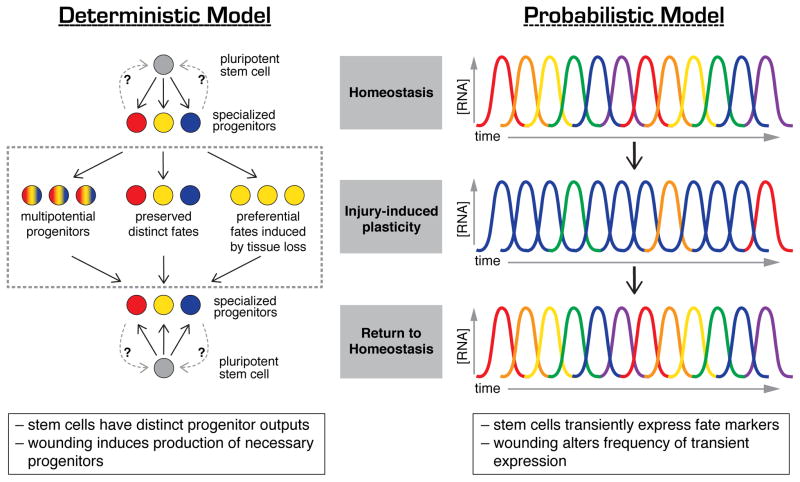
Whether during homeostasis or regeneration, communication between cells in changing environments directly impacts their behavior and output. Considering the large number of neoblasts in planarians, the extensive heterogeneity in expression of cell fate determinants in these cells [36,40,41] and the relative scarcity of cNeoblasts as assayed by single cell transplantation [29], an important question arises: how are stem cells maintained in these changing conditions? Two distinct and testable models become apparent. First, a standard model, in which cNeoblasts represent a core of naïve, pluripotent stem cells constantly undergoing self-renewal (Figure 4, Deterministic Model). However, given that in serial transplantations an estimated 168 neoblasts [76] and only 7/120 single cell injections [29] are capable of rescuing lethally irradiated animals, combined with the uncertainty about the abundance and distribution of cNeoblasts within the animal, it is difficult to explain how small fragments can regenerate complete animals. A second, non-standard model in which cNeoblasts arise stochastically, or on demand, from a larger population of fate-restricted or primed neoblasts (Figure 4, Probabilistic Model). In this latter model, self-renewal becomes a conceptual property not possessed by a discrete population, but transiently held by a small number of cells and arising probabilistically depending on the demands of the animal. If these stem cells stochastically express progenitor markers for specific organs, perhaps injury induces changes in the frequency or periodicity of expression, resulting in altered differentiation of stem cell progeny. Such a model allows us to frame the remarkable plasticity of planarian in terms of dynamic cell states rather than statically defined cell types.
Figure 4.
Two models for progenitor dynamics during homeostasis and regeneration. In the deterministic model, stem cells produce specialized progenitors that statically express specific fate markers. Injury stimulates changes in the rates and types of progenitors produced, allowing animals to respond to different wounds. In the probabilistic model, stem cells and their progenitors transiently express fate markers (RNA concentration represented by curves, with colors representing different genes being expressed at different times). Wounding may alter the frequency and/or persistence of expression of fate determinants, thus producing the necessary cell types required for regeneration.
Our ability to identify these dynamic states of a cell’s life history may have been limited by the methods we have used to probe the system – fixed analysis of tissues and single-timepoint analysis of transcriptomes. With technologies that permit the observation of cell behavior and transcriptional output over time, we are beginning to realize that cell types are not static: even though anatomical position and cell function may be fixed, stochastic and transient changes may occur at the cellular level. As technologies advance, our understanding of the responses of cells to various environmental stimuli will only improve. Future efforts aimed at understanding the dynamics of these cellular states – both in planarians and in other animals – may allow us to redefine cells as having dynamic “states” instead of just fates.
Concluding Remarks: Sic Transit Gloria Cella
Understanding how stem cell behavior is influenced by environmental signals may be the key to deciphering regenerative capacity. Planarians provide intriguing examples of the dynamism of transcriptional activity in differentiated and undifferentiated cells during both tissue homeostasis and injury. However, evidence for the ability of cells to modulate their states under homeostasis and tissue repair is also provided by mammals, particularly by committed intestinal cells that revert to stem cells upon crypt damage. Thus, even though these cells express fate determinants like Dll-1 [12], they can nonetheless be recalled to the stem cell compartment when needed. Another example of plasticity comes from committed enteroendocrine cells expressing fate determinants such as neurogenin 3, which can produce both non-endocrine and non-secretory cells [96,97].
The planarian stem cell population exhibits three key features: 1) the ability to self-renew; 2) the presence of truly pluripotent cells (cNeoblasts); and 3) a high degree of heterogeneity. Although the transcriptional heterogeneity observed appears to be static, current, static experimental methods prohibit the possibility of observing more dynamic transcriptional events. Stochastic and transient transcriptional changes may allow for rapid responses to injury and regeneration demands. Understanding how this may be accomplished will require molecular identification of the cNeoblasts, further characterization of the lineage produced by them during organ regeneration, and additional knowledge about the molecules responsible for communication between stem cells and their environment (Outstanding Questions Box).
Given the desire to harness and manipulate our own cells for the benefit of regenerative medicine, it is essential to understand the complex processes of stem cell maintenance and fate dynamics in different contexts. Studying how planarian stem cells and differentiated cells respond to injury may unlock the key to controlling cell states, allowing us not only to shed light on the mechanisms controlling developmental timing but also on viable approaches leading to the production of particular organs on demand.
Outstanding Questions Box.
What is the wound signal that stimulates distinct stem cell behaviors?
Injury triggers rapid transcriptional changes, cell cycle entry, and repatterning. The signals responsible for initiating these responses are presently unknown.
What distinguishes cNeoblasts from the rest of the stem cell population?
cNeoblast is an operational definition for planarian pluripotent stem cells, but specific. molecular markers for these cells have not yet been identified. Defining specific molecular attributes for these cells will facilitate the identification of their anatomical location and allow analysis of their behavior during regeneration.
How do stem cells sense missing tissues?
Regeneration proceeds without error after any type of amputation, suggesting that stem cells rapidly alter their homeostatic states in response to missing tissue. With their complex anatomy and numerous cell types, how this response is coordinated and executed at the molecular and cellular levels is unclear.
What cellular signaling machinery is present on stem cells?
Extracellular signaling likely instructs changes in cellular dynamics. Our knowledge of the molecules responsible for changes in stem cell behavior is limited, but will continue to expand with high-resolution sequencing and improved molecular techniques.
When does regeneration stop?
Once organs reach their final size and are properly integrated, the accelerated activity of stem cells should subside and return to a homeostatic state. Future work may identify how this is coordinated in the animal.
Trends Box.
Injury induces dramatic changes in tissues. Emerging data from mammalian systems suggest that injury triggers transcriptional and cellular changes in tissues, demonstrating that plasticity of cellular states may be a unique state that facilitates regeneration.
Planarian flatworms readily regenerate after any amputation, demonstrating a rapid response to tissue loss. Because regeneration depends on pluripotent stem cells, swift homeostatic state changes of these cells must occur.
The planarian stem cell population exhibits transcriptional heterogeneity. Many transcription factors required for organ-specific regeneration are expressed in distinct segments of the stem cell population.
After injury, differentiated cells and stem cells alter their transcriptional profiles and cellular behavior. These dynamic changes may describe unique cellular states that drive regeneration.
Acknowledgments
We thank Kim Tu, Beth Duncan and Hanh Thi-Kim Vu for critical reading of this manuscript. And apologize to authors whose work could not be cited due to journal-imposed space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mikkers H, Frisén J. Deconstructing stemness. Embo J. 2005;24:2715–2719. doi: 10.1038/sj.emboj.7600749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanpain C, Fuchs E. Plasticity of epithelial stem cells in tissue regeneration. Science. 2014 doi: 10.1126/science.1242281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donati G, Watt FM. Stem Cell Heterogeneity and Plasticity in Epithelia. Cell Stem Cell. 2015;16:465–476. doi: 10.1016/j.stem.2015.04.014. [DOI] [PubMed] [Google Scholar]
- 4.Lu CP, et al. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell. 2012;150:136–150. doi: 10.1016/j.cell.2012.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Keymeulen A, et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. PNAS. 2008;105:20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xin L, et al. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25:2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 8.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 9.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 10.Clevers H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 11.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 12.van Es JH, et al. Dll1+ secretory progenitor cells revert to stem cells upon crypt damage. Nature Cell Biology. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo W, et al. p63+Krt5+ distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chubb JR, Liverpool TB. Bursts and pulses: insights from single cell studies into transcriptional mechanisms. Curr Opin Genet Dev. 2010;20:478–484. doi: 10.1016/j.gde.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Levine JH, et al. Functional Roles of Pulsing in Genetic Circuits. Science. 2013;342:1188–1193. doi: 10.1126/science.1239999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonev B, et al. MicroRNA-9 Modulates Hes1 Ultradian Oscillations by Forming a Double-Negative Feedback Loop. CellReports. 2012;2:10–18. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodfellow M, et al. microRNA input into a neural ultradian oscillator controls emergence and timing of alternative cell states. Nature Communications. 2014;5:1–10. doi: 10.1038/ncomms4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buettner F, et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat Biotechnol. 2015;33:155–160. doi: 10.1038/nbt.3102. [DOI] [PubMed] [Google Scholar]
- 20.Newmark P, Sánchez Alvarado A. Not your father’s planarian: a classic model enters the era of functional genomics. Nature Review Genetics. 2002;3:210–219. doi: 10.1038/nrg759. [DOI] [PubMed] [Google Scholar]
- 21.Cebrià F, et al. The expression of neural-specific genes reveals the structural and molecular complexity of the planarian central nervous system. Mech Develop. 2002;116:199–204. doi: 10.1016/s0925-4773(02)00134-x. [DOI] [PubMed] [Google Scholar]
- 22.Okamoto K, et al. Neural projections in planarian brain revealed by fluorescent dye tracing. Zoological Science. 2005;22:535–546. doi: 10.2108/zsj.22.535. [DOI] [PubMed] [Google Scholar]
- 23.Lapan SW, Reddien PW. dlx and sp6-9 Control Optic Cup Regeneration in a Prototypic Eye. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsthoefel DJ, et al. Stem cell-based growth, regeneration, and remodeling of the planarian intestine. Developmental Biology. 2011;356:445–459. doi: 10.1016/j.ydbio.2011.05.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scimone ML, et al. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rink JC, et al. The maintenance and regeneration of the planarian excretory system are regulated by EGFR signaling. Development. 2011;138:3769–3780. doi: 10.1242/dev.066852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newmark P, Sánchez Alvarado A. Bromodeoxyuridine specifically labels the regenerative stem cells of planarians. Developmental Biology. 2000;220:142–153. doi: 10.1006/dbio.2000.9645. [DOI] [PubMed] [Google Scholar]
- 28.Cowles MW, et al. Genome-wide analysis of the bHLH gene family in planarians identifies factors required for adult neurogenesis and neuronal regeneration. Development. 2013;140:4691–4702. doi: 10.1242/dev.098616. [DOI] [PubMed] [Google Scholar]
- 29.Wagner DE, et al. Clonogenic Neoblasts Are Pluripotent Adult Stem Cells That Underlie Planarian Regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solana J, et al. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNA-seq, RNA interference and irradiation approach. Genome Biol. 2012 doi: 10.1186/gb-2012-13-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner DE, et al. Genetic Regulators of a Pluripotent Adult Stem Cell System in Planarians Identified by RNAi and Clonal Analysis. Cell Stem Cell. 2012;10:299–311. doi: 10.1016/j.stem.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baguñà J. The planarian neoblast: the rambling history of its origin and some current black boxes. Int J Dev Biol. 2012;52:19–37. doi: 10.1387/ijdb.113463jb. [DOI] [PubMed] [Google Scholar]
- 33.Rink JC. Stem cell systems and regeneration in planaria. Dev Genes Evol. 2012;223:67–84. doi: 10.1007/s00427-012-0426-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayashi T, et al. Single-cell gene profiling of planarian stem cells using fluorescent activated cell sorting and its “index sorting” function for stem cell research. Dev Growth Differ. 2010;52:131–144. doi: 10.1111/j.1440-169X.2009.01157.x. [DOI] [PubMed] [Google Scholar]
- 35.Shibata N, et al. Comprehensive gene expression analyses in pluripotent stem cells of a planarian, Dugesia japonica. Int J Dev Biol. 2012;56:93–102. doi: 10.1387/ijdb.113434ns. [DOI] [PubMed] [Google Scholar]
- 36.van Wolfswinkel JC, et al. Single-Cell Analysis Reveals Functionally Distinct Classes within the Planarian Stem Cell Compartment. Cell Stem Cell. 2014;15:326–339. doi: 10.1016/j.stem.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pearson BJ, Sánchez Alvarado A. A planarian p53 homolog regulates proliferation and self-renewal in adult stem cell lineages. Development. 2010;137:213–221. doi: 10.1242/dev.044297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lapan SW, Reddien PW. Transcriptome Analysis of the Planarian Eye Identifies ovo as a Specific Regulator of Eye Regeneration. CellReports. 2012;2:294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsthoefel DJ, et al. An RNAi Screen Reveals Intestinal Regulators of Branching Morphogenesis, Differentiation, and Stem Cell Proliferation in Planarians. Dev Cell. 2012;23:691–704. doi: 10.1016/j.devcel.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler CE, et al. Selective amputation of the pharynx identifies a FoxA-dependent regeneration program in planaria. eLife. 2014 doi: 10.7554/eLife.02238.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scimone ML, et al. Neoblast Specialization in Regeneration of the Planarian Schmidtea mediterranea. Stem Cell Reports. 2014 doi: 10.1016/j.stemcr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reddien PW. Specialized progenitors and regeneration. Development. 2013;140:951–957. doi: 10.1242/dev.080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reddien P, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 44.Baguñà J, et al. Experimental Embryology in Aquatic Plants and Animals. Springer; US: 1990. Growth, degrowth and regeneration as developmental phenomena in adult freshwater planarians; pp. 129–162. [Google Scholar]
- 45.Kang H, Alvarado A. Flow cytometry methods for the study of cell-cycle parameters of planarian stem cells. Dev Dyn. 2009 doi: 10.1002/dvdy.21928. [DOI] [PubMed] [Google Scholar]
- 46.Pellettieri J, et al. Cell death and tissue remodeling in planarian regeneration. Developmental Biology. 2010;338:76–85. doi: 10.1016/j.ydbio.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oviedo N, et al. Allometric scaling and proportion regulation in the freshwater planarian Schmidtea mediterranea. Dev Dyn. 2003;226:326–333. doi: 10.1002/dvdy.10228. [DOI] [PubMed] [Google Scholar]
- 48G.onzález-Estévez C, et al. Decreased neoblast progeny and increased cell death during starvation-induced planarian degrowth. Int J Dev Biol. 2012;56:83–91. doi: 10.1387/ijdb.113452cg. [DOI] [PubMed] [Google Scholar]
- 49.Takeda H, et al. Planarians maintain a constant ratio of different cell types during changes in body size by using the stem cell system. Zoological Science. 2009;26:805–813. doi: 10.2108/zsj.26.805. [DOI] [PubMed] [Google Scholar]
- 50.Abeloos M. Recherches expérimentales sur la croissance et la régénération chez les planaires. Bull Biol. 1930;1:1–140. [Google Scholar]
- 51.Lillie F. Some Notes on Regeneration and Regulation in Planarians. The American Naturalist. 1900;34:173–177. [Google Scholar]
- 52.Schultz E. Über Reduktionen. I. Über Hungerserscheinungen bei Planaria lactea. Arch Entwm. 1904;18:555–577. [Google Scholar]
- 53.Berninger J. Über die Einwirkung des Hungers auf Planarien. J Zool Jahrb. 1911;30:181–216. [Google Scholar]
- 54.Baguñà J, Romero R. Quantitative analysis of cell types during growth, degrowth and regeneration in the planarians Dugesia mediterranea and Dugesia tigrina. Hydrobiologia. 1981;84:181–194. [Google Scholar]
- 55.Cowles MW, et al. COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- 57.Koinuma S, et al. Planaria FoxA (HNF3) homologue is specifically expressed in the pharynx-forming cells. Gene. 2000;259:171–176. doi: 10.1016/s0378-1119(00)00426-1. [DOI] [PubMed] [Google Scholar]
- 58.Morgan T. Regeneration. The Macmillan Company; 1901. [Google Scholar]
- 59.Kobayashi C, et al. The process of pharynx regeneration in planarians. Developmental Biology. 1999;211:27–38. doi: 10.1006/dbio.1999.9291. [DOI] [PubMed] [Google Scholar]
- 60.Bueno D, et al. Maintenance of A/P body regions in planarians by TCEN49, a putative cystine-knot neurotrophin. Belg J Zool. 2001;131:89–95. [Google Scholar]
- 61.Bueno D, et al. Regeneration of gross molecular body regions in planarian: from molecules to organs. Int J Dev Biol. 2001;45:S121–S122. [Google Scholar]
- 62.Reddien P, et al. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- 63.Gurley KA, et al. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Developmental Biology. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wenemoser D, et al. A molecular wound response program associated with regeneration initiation in planarians. Genes Dev. 2012;26:988–1002. doi: 10.1101/gad.187377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandmann T, et al. The head-regeneration transcriptome of the planarian Schmidtea mediterranea. Genome Biol. 2011 doi: 10.1186/gb-2011-12-8-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends Genet. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petersen CP, Reddien PW. A wound-induced Wnt expression program controls planarian regeneration polarity. PNAS. 2009;106:17061–17066. doi: 10.1073/pnas.0906823106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petersen CP, Reddien PW. Polarized notum Activation at Wounds Inhibits Wnt Function to Promote Planarian Head Regeneration. Science. 2011;332:852–855. doi: 10.1126/science.1202143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kato K, et al. Dorsal and ventral positional cues required for the onset of planarian regeneration may reside in differentiated cells. Developmental Biology. 2001;233:109–121. doi: 10.1006/dbio.2001.0226. [DOI] [PubMed] [Google Scholar]
- 70.Saló E, Baguñà J. Cell movement in intact and regenerating planarians. Quantitation using chromosomal, nuclear and cytoplasmic markers. J Embryol Exp Morphol. 1985;89:57–70. [PubMed] [Google Scholar]
- 71.Witchley JN, et al. Muscle Cells Provide Instructions for Planarian Regeneration. CellReports. 2013 doi: 10.1016/j.celrep.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reuter H, et al. β-Catenin-Dependent Control of Positional Information along the AP Body Axis in Planarians Involves a Teashirt Family Member. CellReports. 2015;10:253–265. doi: 10.1016/j.celrep.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 73.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Developmental Biology. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. I. Mitotic studies during growth, feeding and starvation. Journal of Experimental Zoölogy. 1976;195:53–64. [Google Scholar]
- 75.Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n.sp. II. Mitotic studies during regeneration, and a possible mechanism of blastema formation. Journal of Experimental Zoölogy. 1976;195:65–80. [Google Scholar]
- 76.Guedelhoefer OC, Sánchez Alvarado A. Amputation induces stem cell mobilization to sites of injury during planarian regeneration. Development. 2012;139:3510–3520. doi: 10.1242/dev.082099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Scimone ML, et al. A forkhead Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1003999.s007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogg MC, et al. Stem cell-dependent formation of a functional anterior regeneration pole in planarians requires Zic and Forkhead transcription factors. Developmental Biology. 2014;390:136–148. doi: 10.1016/j.ydbio.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 79.Vásquez-Doorman C, Petersen CP. zic-1 Expression in Planarian Neoblasts after Injury Controls Anterior Pole Regeneration. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004452.s013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roberts-Galbraith RH, Newmark PA. Follistatin antagonizes activin signaling and acts with notum to direct planarian head regeneration. PNAS. 2013;110:1363–1368. doi: 10.1073/pnas.1214053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Blassberg RA, et al. PBX/extradenticle is required to re-establish axial structures and polarity during planarian regeneration. Development. 2013;140:730–739. doi: 10.1242/dev.082982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fraguas S, et al. EGFR signaling regulates cell proliferation, differentiation and morphogenesis during planarian regeneration and homeostasis. Developmental Biology. 2011;354:87–101. doi: 10.1016/j.ydbio.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 83.Sakurai T, et al. The planarian P2X homolog in the regulation of asexual reproduction. Int J Dev Biol. 2012;56:173–182. doi: 10.1387/ijdb.113439ts. [DOI] [PubMed] [Google Scholar]
- 84.Gaviño MA, et al. Tissue absence initiates regeneration through Follistatin-mediated inhibition of Activin signaling. eLife. 2013 doi: 10.7554/eLife.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tu KC, et al. TORC1 is required to balance cell proliferation and cell death in planarians. Developmental Biology. 2012;365:458–469. doi: 10.1016/j.ydbio.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Almuedo-Castillo M, et al. JNK Controls the Onset of Mitosis in Planarian Stem Cells and Triggers Apoptotic Cell Death Required for Regeneration and Remodeling. PLoS Genet. 2014 doi: 10.1371/journal.pgen.1004400.s010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Day SJ, Lawrence PA. Measuring dimensions: the regulation of size and shape. Development. 2000;127:2977–2987. doi: 10.1242/dev.127.14.2977. [DOI] [PubMed] [Google Scholar]
- 88.Lander Arthur D, et al. Cell Lineages and the Logic of Proliferative Control. PLoS Biol. 2009 doi: 10.1371/journal.pbio. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Stanger BZ. The biology of organ size determination. Diabetes, Obesity and Metabolism. 2008;10:16–22. doi: 10.1111/j.1463-1326.2008.00938.x. [DOI] [PubMed] [Google Scholar]
- 90.Bullough WS. Mitotic and Functional Homeostasis: A Speculative Review. Cancer Research. 1965;25:1683–2577. [PubMed] [Google Scholar]
- 91.McPherron AC, et al. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 1997;387:83–90. doi: 10.1038/387083a0. [DOI] [PubMed] [Google Scholar]
- 92.McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. PNAS. 1997;94:12457–12461. doi: 10.1073/pnas.94.23.12457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ziller-Sengel C. Sur le facteur inhibiteur de la régénération du pharynx chez les planaires d’eau douce. Bul Soc Zool France. 1967;92:295–302. [Google Scholar]
- 94.Ziller-Sengel C. Investigation of the inhibition of the regeneration of the planarian pharynx. I. Evidence of an auto-inhibiting factor found in the regenerating pharynx. Journal of Embryology and Experimental Morphology. 1967;18:91–105. [PubMed] [Google Scholar]
- 95.Ziller-Sengel C. Investigation of the inhibition of the regeneration of the planarian pharynx. II. Variations of the amount of the inhibitor dependent on the species and the phase of regeneration. Journal of Embryology and Experimental Morphology. 1967;18:107–119. [PubMed] [Google Scholar]
- 96.Jenny M, et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. Embo J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schonhoff SE, et al. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Developmental Biology. 2004;270:443–454. doi: 10.1016/j.ydbio.2004.03.013. [DOI] [PubMed] [Google Scholar]