Abstract
The present study establishes a visualization method for the measurement of the distribution and localization of protein/peptide constituents within a single poly-lactide-co-glycolide (PLGA) microsphere using synchrotron radiation–based Fourier-transform infrared spectromicroscopy (SR-FTIR). The representative infrared wavenumbers specific for protein/peptide (Exenatide) and excipient (PLGA) were identified and chemical maps at the single microsphere level were generated by measuring and plotting the intensity of these specific bands. For quantitative analysis of the distribution within microspheres, Matlab software was used to transform the map file into a 3D matrix and the matrix values specific for the drug and excipient were extracted. Comparison of the normalized SR-FTIR maps of PLGA and Exenatide indicated that PLGA was uniformly distributed, while Exenatide was relatively non-uniformly distributed in the microspheres. In conclusion, SR-FTIR is a rapid, nondestructive and sensitive detection technology to provide the distribution of chemical constituents and functional groups in microparticles and microspheres.
KEY WORDS: Fourier-transform infrared spectromicroscopy, Protein distribution, Microsphere, Exenatide, PLGA
Graphical abstract
A visualization method is investigated for the measurement of the distribution and localization of protein/peptide within single microsphere by SR-FTIR. Chemical maps at the single microsphere level were generated by the intensity of specific infrared bands. The relative infrared intensity ratios and optical path normalization provided internal material distribution.
1. Introduction
Protein/peptide drugs usually have high specificity and high activity with low dosage. The microparticles or microspheres are designed to deliver proteins for enhanced absorption and bioavailability with modified release1, 2. With the progress of science and technology, the development of microparticles gradually moves towards accurate control of their structures3, 4.
There are many parameters to the release kinetics of proteins from microspheres, e.g., polymer nature, morphology, structures and drug distribution, to which drug distribution in microspheres is essential5. A well investigated distribution of drug in a single microsphere will help deepen the knowledge in design and optimization for the formulation and performance of microspheres. Recent research has studied the relationship of particle structure and drug release from single pellets using synchrotron radiation X-ray computed microtomography (SR-μCT) combined with a sensitive liquid chromatography–tandem mass spectrometry (LC/MS/MS) method. It was found that the dissolution rate of the individual particles correlated with the combined influences of drug loading, volume and surface area of the pellets, and the void microstructures within the pellet was critical during drug release6.
At present, the characterization methods for the structures of microspheres are conventional light microscopy (LM)7, scanning electron microscopy (SEM)8, traditional infrared spectroscopy and confocal laser scanning microscopy (CLSM)9, 10, 11, 12, 13. LM is capable of determining the shape and size of particles, but this method is impeded by the scattering or emission of light from structures outside the optical focal plane, which reduces the quality of the images. For microspheres prepared with materials with low transmittance or opacity, only their morphology and outer structures can be observed. SEM can provide extremely high spatial resolution, but the penetration ability is not strong. It usually requires a relatively complex sample pre-treatment and it is unable visualize the distribution of materials within the microspheres14. To identify and localize the encapsulated drug or the distribution of the involved polymers is essential for the characterization of the microspheres. Traditional infrared spectroscopy cannot reflect the differences in the composition and distribution of the objects even though it can identify the components of the microspheres, but the component measured by traditional infrared spectroscopy is the average composition of the samples, as the microspheres are very small. In addition, the distribution of each component in the microspheres is affected by the pH of medium and other environmental factors. CLSM has already been used in the evaluation and characterization of solid pharmaceutical formulations, including the release kinetics of the entrapped drugs9, 10, localization of the oil phase11, characterization of the entrapped phase and polymer structures on the surface12 and the examination of the permeability of microparticles by using fluorescein-containing aqueous media13. CLSM permits the identification of several compounds through the use of different fluorescence labels, but the labeling process is cumbersome.
Synchrotron radiation based on Fourier-transform infrared spectromicroscopy (SR-FTIR) can analyze samples at the micron level or micro-sample areas15, 16. SR-FTIR has been employed in the characterization of structure and function of stain-free and label-free samples. As a non-destructive approach, SR-FTIR microscopy can keep the crystal structure and morphology of the samples with advantages of synchrotron light brightness (which is usually 100–1000 times brighter than a conventional globar source and has a small effective source size), and is capable of exploring the molecular chemistry within the microstructures of samples with a high signal-to-noise ratio (S/N) at ultraspatial resolutions17. This technique is able to provide information including the quantity, composition, structure and distribution of chemical constituents and functional groups in samples and extract more detailed structural information. The technique has recently been utilized to monitor the changing chemistry in a single living cell18 and in vitro for cell uptake studies of drugs19 by quantitative analysis of the protein, nucleic acid and phospholipid components20, 21.
In this report we have developed a mathematical post-test data processing procedure to overcome the difficulties in mapping and analyzing globularly structured microspheres and demonstrate its value by measurement of the distribution of Exenatide and PLGA in the microspheres via SR-FTIR.
2. Materials and methods
2.1. Materials
Exenatide was a kind gift from Jilin University (China). PLGA (LA/GA=75:25) was purchased from Lakeshore Biomaterials (USA). All other chemicals were of analytical grade.
The microspheres were prepared using the W/O/W emulsion-solvent evaporation technique22. Briefly, the Exenatide aqueous solution was added into 0.9 mL dichloromethane solution under stirring with a homogenizer at 5000 rpm to prepare a water-in-oil primary emulsion. This emulsion was added to 10 mL of water containing 1% (w/w) polyvinyl alcohol (PVA) and 5% (w/w) sodium chloride under stirring at 1000 rpm for 2 min. Finally, the solidified microspheres were washed three times using distilled water and then freeze-dried before storage. The diameters of microsphere were about 50 μm.
2.2. SR-FTIR spectromicroscopy
Synchrotron radiation from a bending magnet was collected, collimated and transported to a commercial FTIR interferometer bench. After modulation by the interferometer, a commercial infrared microscope focused the beam onto the sample using all-reflecting optics. The sample was placed on the sample stage and the sample stage position was controlled by a computer. The reflected light from the sample was collected by the microscope optics and sent to an IR detector. A computer performed a Fourier transform on the measured interferogram to obtain an infrared spectrum for each sample location23. To characterize the distribution of each composition in the microspheres, SR-FTIR spectromicroscopy was performed with the BL01B beam line at SSRF24 (Shanghai Synchrotron Radiation Facility). Spectra were recorded on a Nicolet Continuμm XL microscope (Thermo Fisher Scientific) equipped with a 250×250 μm2 liquid nitrogen cooled MCT/A detector, a 32X/NA0.65 Schwarzschild objective, a motorized knife-edge aperture, and a Prior XYZ motorized stage and coupled with Nicolet 6700 spectrometer (Thermo Fisher) equipped with a Michelson interferometer. Spectra of Exenatide and PLGA were recorded in transmission mode between 650 cm−1 and 4000 cm−1, with 128 scans at 8 cm−1 resolution, 10 μm×10 μm aperture dimension on the BaF2 substrate. At least three microspheres were mapped in transmission mode between 650 cm−1 and 4000 cm−1, with 64 scans at 8 cm−1 resolution, 10 μm×10 μm aperture dimension and 5 μm×5 μm step size. The background was collected through a blank substrate.
2.3. Data and image processing
With the Ominic software (Thermo Fisher), chemical maps were created and analyzed after baseline corrections were applied to each spectrum. The following spectral bands were measured: PLGA, 1770–1740 cm−1 (C=O bond); Exenatide, 1670–1625 cm−1 (amide bands). Further analysis of the maps has been carried out with Matlab 2012a. The two dimensional (2D) matrix at a specific wavenumber was extracted from the map file. For accuracy a bilinear interpolation algorithm was applied. The outline of the microspheres was specified based on a contour line and fitted with a model sphere to determine the center coordinates, yielding the radius of the optical path of each point. Then the absorbance value at each point was normalized by the optical path. For investigation of the relative distribution of Exenatide compared to PLGA, the image at specific wavenumber of Exenatide was divided by that of PLGA.
3. Results and discussion
3.1. Specific band analysis
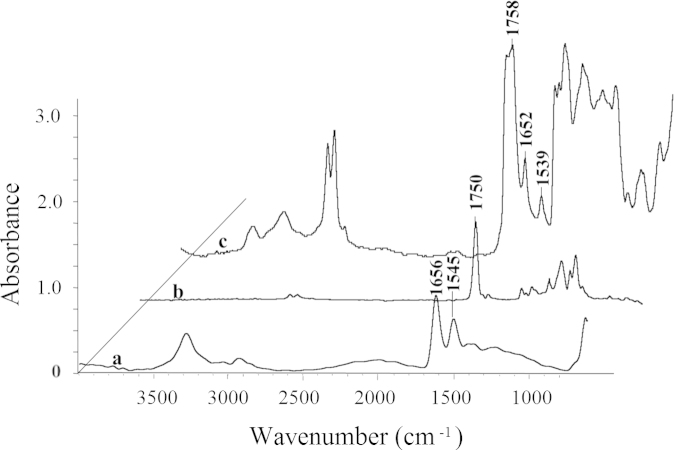
Using IR microspectroscopy to probe Exenatide distribution in the microsphere, the spectra of Exenatide and matrix PLGA were analyzed for the determination of unique features by which the two components could be distinguished. The PLGA particle is very large for direct spectra analysis, and it is also difficult to grind. When collecting the spectra of PLGA, the PLGA was dissolved in acetone first, and 2 μL of solution was dropped on the BaF2 substrate for drying. The spectra of Exenatide were collected also with the same treatment. Fig. 1 illustrates the FTIR microspectra of (a) Exenatide, (b) PLGA and (c) a single microsphere. The IR spectrum of Exenatide is dominated by the amide I and amide II bands, centered at 1656 and 1545 cm−1, respectively. The amide I band is due to the stretching vibrations of the C=O bond in the backbone of the protein. The amide II band arises from a combination of C–N stretching and N–H bending vibrations of the protein backbone25. For the microsphere matrix, C=O bond of PLGA absorbed at 1750 cm−1, which is different from the absorbance features of Exenatide. The absorption at 1450, 1380, 1270 and 1190 cm−1 is attributed to CH2 and CH3 wagging and deformation of PLGA. It is important to note that the specific bands of both PLGA and Exenatide can be identified from the absorbance spectra of the microsphere without significant interference between each other.
Figure 1.
SR-FTIR of (a) Exenatide, (b) PLGA and (c) single microsphere.
3.2. Analysis of the mapping image
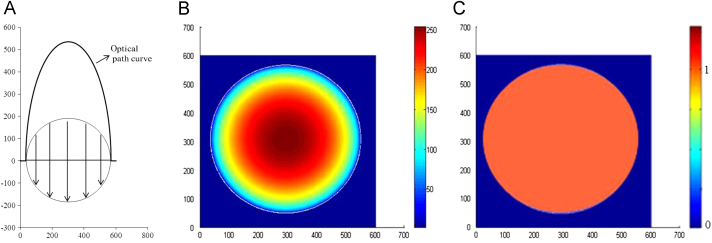
Microspheres composed mainly of Exenatide and PLGA were analyzed in this paper. On the basis of the different spectrums between Exenatide and the microsphere matrix map, the distribution of protein in the microsphere was determined. The peaks at 1750 cm−1 (PLGA carbonyl ester bond absorption), 1656 cm−1 (amide I) and 1545 cm−1 (amide II) were selected to generate the chemical imaging which can reflect the intensity distribution of the different compositions in 2D space. Fig. 2 shows a 2D image of standard sphere model in which the material distribution is uniform. As the sample is spherical, the optical path of the light through microsphere is different from point to point as shown in Fig. 2A, so the apparent observed distribution of a homogeneous in microsphere that directly read from the map is shown in Fig. 2B.
Figure 2.
2D image of standard sphere. (A) Optical path length and optical path curve; (B) the uniform material distribution in a sphere read as nonuniform 2D image by SR-FTIR mapping; (C) the uniform distribution intensity of a typical homogeneous sphere was recovered after being normalized with the length of optical path.
3.2.1. Analysis of chemical maps
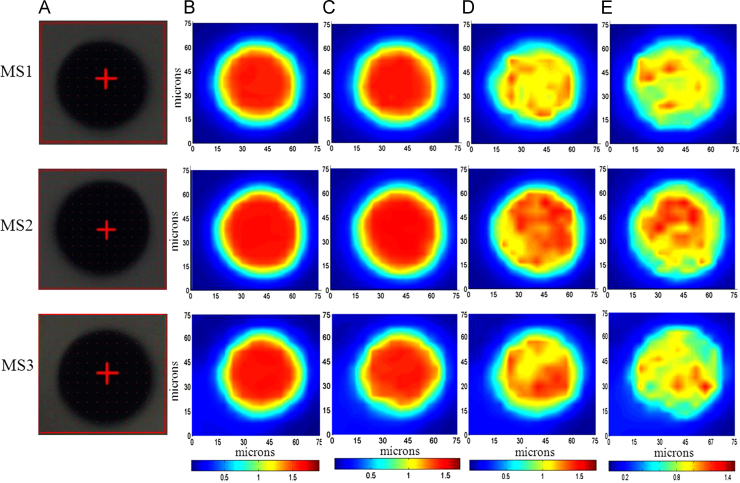
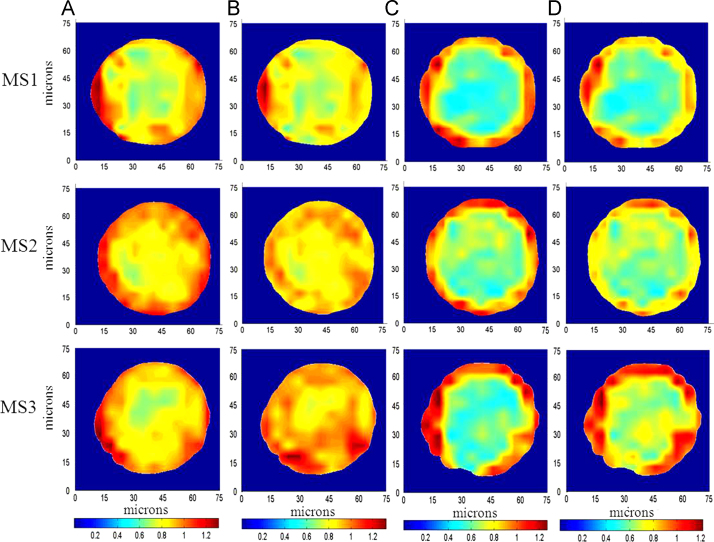
The microspheres mounted on a BaF2 substrate were mapped using SR-FTIR and the local SR-FTIR spectra were recorded at the single microsphere level. Chemical maps at the single microsphere level were generated by measuring and plotting the intensity of a specific band for Exenatide (amide I, amide II) and PLGA (carbonyl ester bond). Maps were plotted using the absorption spectra (maximum absorption is represented in red, and minimum absorption is represented in blue). Fig. 3 shows the images of three different microspheres. The maximal intensities of amide bond and PLGA plot allowed us to recognize the distribution of Exenatide and PLGA.
Figure 3.
Chemical imaging using SR-FTIR of three single microspheres (MS1–MS3). The red boxes indicate the area imaged by the IR microscope. (A) Light microscope images of PLGA microspheres; (B–E) chemical imaging for the presence of PLGA (carbonyl ester bond), PLGA (CH2 wagging), proteins (amide I) and proteins (amide II) are shown in the single microsphere.
Fig. 3B and C demonstrates chemical imaging for the presence of PLGA. These particles have strong absorption and high uniformity in the center of the microspheres. Compared with Fig. 2B, the distribution of PLGA is highly similar, indicating that the distribution of PLGA in the microspheres is quite uniform. For the chemical imaging of Exenatide (Fig. 3D and E), the absorption intensity is different from that of the PLGA. Furthermore, the nonuniform image density demonstrates that the distribution of Exenatide is not uniform throughout this microsphere. The distribution of PLGA in the different microspheres (MS1–MS3) is similar, and all are relative uniform. However, the distribution of Exenatide in different microspheres (MS1–MS3) shows significant differences. The absorption intensity of Exenatide in different microspheres (MS1–MS3) is also different due to the different amounts of protein content in different microspheres.
3.2.2. Optical path normalization
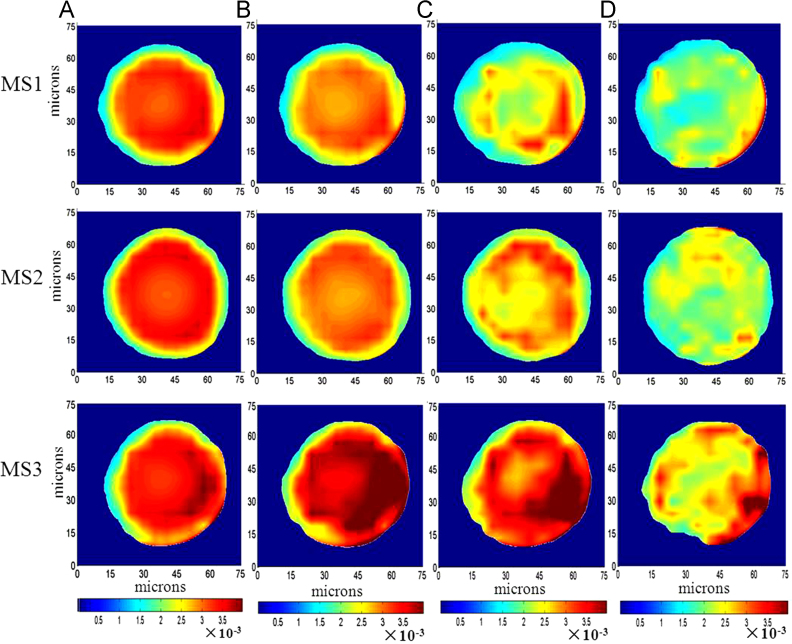
Compared with the standard microsphere, it is reasonable to suppose that the distribution of PLGA in the microspheres relatively uniform. In order to understand the distribution of Exenatide and PLGA further, we normalized the optical path of the maps at PLGA-specific bonds to reduce the distortion introduced by the 3D shape of the microspheres. First, the microsphere was considered as a model sphere. Then, the absorbance value of each point was divided by the optical path. The value of the normalized optical path represented the substance content at each pixel area. The values that are same indicated uniform distribution. The values that are not consistent represented uneven distribution. Fig. 4 shows the images of optical path normalization of different microspheres. In this study, the maps were collected with 10 μm×10 μm aperture dimension and 5 μm step size. The absorbance intensity at the area of 5 μm×5 μm actually scanned with a beam as 10 μm×10 μm. As the edge area was smaller than the aperture, the absorbance intensity for the part outside of the edge was gradually increased, while the part inside of the edge was decreased. Therefore, the normalized intensity of PLGA at the margin area was lower than that of the centric area. The size and the morphology could dramatically alter the infrared spectra26, 27. The absorption close to the edge was distorted by Mie-type scattering, which resulted in weak absorption for the edge region of the microsphere. For the normalized image at peaks of 1750 and 1450 cm−1 (Fig. 4A and B), the relatively weak absorption in the center indicated that the microsphere was internally porous.
Figure 4.
Optical path normalized image of different microspheres (MS1–MS3). (A) Carbonyl ester bond at 1750 cm−1 of PLGA; (B) CH2 wagging at 1450 cm−1 of PLGA; (C) amide I bond at 1656 cm−1; (D) amide II bond at 1545 cm−1.
3.2.3. Distribution of relative intensity ratios
SR-FTIR microspectroscopy allows us to generate 2D chemical images by acquiring infrared spectra at each pixel of the desired map. Thus, by monitoring the relative intensity ratios of amide bonds (1650 and 1540 cm−1) to ester bond (1750 cm−1) of PLGA, we can describe the samples in terms of relative protein distribution. The distribution of ester bond/amide band relative intensity ratios calculated for SR-FTIR is shown as a map in Fig. 5. The red and the blue represent relatively strong and weak regions, respectively. The relative intensity is more consistent in the MS2 and MS3 compared with MS1. We conclude that the distribution of Exenatide in the microspheres (MS2 and MS3) is largely uniform. For MS1, as the intensity ratio is weak in the center and not uniform, we conclude that the distribution of Exenatide is nonuniform. As indicated from the results of image relative intensity ratios, Exenatide has greater variation in distribution in microspheres.
Figure 5.
Spectral images constructed using the relative ratio of amid I bond/carbonyl ester bond of PLGA (A), amide II bond/carbonyl ester bond of PLGA (B), amid I bond/CH2 wagging of PLGA (C), amide II bond/CH2 wagging of PLGA (D) in different microspheres.
4. Conclusions
In this study a visualization method for the distribution and localization of protein/peptide and excipient components within single microsphere using SR-FTIR was established. The representative infrared wavenumbers for protein/peptide and excipient were identified. The results from SR-FTIR showed that the absorbance spectra of Exenatide at 1656 and 1545 cm−1 from the amide I and amide II bonds differed from the PLGA spectrum at 1750 cm−1 due to the C=O bond. The chemical maps at the single microsphere level were generated by measuring and plotting the intensity of specific bands for Exenatide (amide I and amide II) and PLGA (C=O bond). After map analysis and point-by-point optical path normalization, we concluded that the distribution of PLGA was uniform, while the distribution of Exenatide was nonuniform within microspheres. In conclusion, as a nondestructive and sensitive detection technology, SR-FTIR provides the chemical distribution of constituents based on the functional group distribution in the samples. A new procedure was developed to process the data and make it accurate and readable. The study of optical path normalization has demonstrated that the error caused by the shape of particles cannot be ignored. This study has introduced the relative intensity ratio as a new index to provide the chemical distribution after SR-FTIR mapping test.
Acknowledgments
The authors are grateful for the financial support from the National Natural Science Foundation of China (Nos. 81273453 and 81430087).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Min Chen, Email: chenmin@sinap.ac.cn.
Jiwen Zhang, Email: jwzhang@simm.ac.cn.
References
- 1.Tan ML, Choong PFM, Dass CR. Recent developments in liposomes, microparticles and nanoparticles for protein and peptide drug delivery. Peptides. 2010;31:184–193. doi: 10.1016/j.peptides.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Li M, Rouaud O, Poncelet D. Microencapsulation by solvent evaporation: state of the art for process engineering approaches. Int J Pharm. 2008;363:26–39. doi: 10.1016/j.ijpharm.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 3.Pivette P, Faivre V, Mancini L, Gueutin C, Daste G, Ollivon M. Controlled release of a highly hydrophilic API from lipid microspheres obtained by prilling: analysis of drug and water diffusion processes with X-ray-based methods. J Control Release. 2012;158:393–402. doi: 10.1016/j.jconrel.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Mönkäre J, Pajander J, Hakala R A, Savolainen P, Järveläinen M, Korhonen H. Characterization of internal structure, polymer erosion and drug release mechanisms of biodegradable poly(ester anhydride)s by X-ray microtomography. Eur J Pharm Sci. 2012;47:170–178. doi: 10.1016/j.ejps.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Yang YY, Chung TS, Ng NP. Morphology, drug distribution, and in vitro release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion solvent extraction/evaporation method. Biomaterials. 2001;22:231–241. doi: 10.1016/s0142-9612(00)00178-2. [DOI] [PubMed] [Google Scholar]
- 6.Yang S, Yin XZ, Wang CF, Li HY, He Y, Xiao TQ. Release behaviour of single pellets and internal fine 3D structural features co-define the in vitro drug release profile. AAPS J. 2014;16:860–871. doi: 10.1208/s12248-014-9611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J.L., Lu J.F. In vitro drug release profiles and mucoadhesive property of bioadhesive microspheres of metronicazole. Acta Pharm Sin. 2002;37:226–228. [PubMed] [Google Scholar]
- 8.Jelvehgari M, Barar J, Valizadeh H, Shadrou S, Nokhodchi A. Formulation, characterization and in vitro evaluation of theophylline-loaded Eudragit RS 100 microspheres prepared by an emulsion-solvent diffusion/evaporation technique. Pharm Dev Technol. 2011;16:637–644. doi: 10.3109/10837450.2010.508075. [DOI] [PubMed] [Google Scholar]
- 9.Zaky A, Elbakry A, Ehmeret A, Breuning M, Goepferich A. The mechanism of protein release from triglyceride microspheres. J Control Release. 2010;147:202–210. doi: 10.1016/j.jconrel.2010.07.110. [DOI] [PubMed] [Google Scholar]
- 10.De Smedt SC, Meyvis TK, Van Oostveldt P, Demeester J. A new microphotolysis based approach for mapping the mobility of drugs in microscopic drug delivery devices. Pharm Res. 1999;16:1639–1642. doi: 10.1023/a:1011988109989. [DOI] [PubMed] [Google Scholar]
- 11.Lamprecht A, Schäfer UF, Lehr CM. Characterization of microcapsules by confocal laser scanning microscopy: structure, capsule wall composition and encapsulation rate. Eur J Pharm Biopharm. 2000;49:1–9. doi: 10.1016/s0939-6411(99)00063-6. [DOI] [PubMed] [Google Scholar]
- 12.Lamprecht A, Schäfer UF, Lehr CM. Structural analysis of microparticles by confocal laser scanning microscopy. AAPS PharmSciTech. 2000;1:10–19. doi: 10.1208/pt010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou XL, He JT, Zhou ZT, Ma SF, Jiang Y, Wang Y. Effect of NaCl in outer water phase on the characteristics of BSA-loaded PLGA sustained-release microspheres fabricated by a solid-in-oil-in-water emulsion technique. Acta Pharm Sin. 2010;45:1057–1063. [PubMed] [Google Scholar]
- 14.Wang SB, Guo SR. Formation mechanism and release behavior of poly(ε-caprolactone) microspheres containing disodium norcantharidate. Eur J Pharm Biopharm. 2008;69:1176–1181. doi: 10.1016/j.ejpb.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Martinez L, Agnely F, Leclerc B, Siepmann J, Cotte M, Geiger S. Cross-linking of chitosan and chitosan/poly(ethylene oxide) beads: a theoretical treatment. Eur J Pharm Biopharm. 2007;67:339–348. doi: 10.1016/j.ejpb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 16.Marmorato P, Ceccone G, Gianoncelli A, Pascolo L, Ponti J, Rossi F. Cellular distribution and degradation of cobalt ferrite nanoparticles in Balb/3T3 mouse fibroblasts. Toxicol Lett. 2011;207:128–136. doi: 10.1016/j.toxlet.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 17.Pascolo L, Bortot B, Benseny-Cases N, Gianoncelli A, Tosi G, Ruozi B. Detection of PLGA-based nanoparticles at a single-cell level by synchrotron radiation FTIR spectromicroscopy and correlation with X-ray fluorescence microscopy. Int J Nanomed. 2014;9:2791–2801. doi: 10.2147/IJN.S58685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holman HY, Miles R, Hao Z, Wozei E, Anderson LM, Yang H. Real-time chemical imaging of bacterial activity in biofilms using open-channel microfluidics and synchrotron FTIR spectromicroscopy. Anal Chem. 2009;81:8564–8570. doi: 10.1021/ac9015424. [DOI] [PubMed] [Google Scholar]
- 19.Clède S, Lambert F, Sandt C, Gueroui Z, Delsuc N, Dumas P. Synchrotron radiation FTIR detection of a metal-carbonyl tamoxifen analog. Correlation with luminescence microscopy to study its subcellular distribution. Biotechnol Adv. 2013;31:393–395. doi: 10.1016/j.biotechadv.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 20.Kastyak-Ibrahim MZ, Nasse MJ, Rak M, Hirschmugl C, Del Bigio MR, Albensi BC. Biochemical label-free tissue imaging with subcellular-resolution synchrotron FTIR with focal plane array detector. Neuroimage. 2012;60:376–383. doi: 10.1016/j.neuroimage.2011.11.069. [DOI] [PubMed] [Google Scholar]
- 21.Jamin N, Miller L, Moncuit J, Fridman WH, Dumas P, Teillaud JL. Chemical heterogeneity in cell death: combined synchrotron IR and fluorescence microscopy studies of single apoptotic and necrotic cells. Biopolymers. 2003;72:366–373. doi: 10.1002/bip.10435. [DOI] [PubMed] [Google Scholar]
- 22.Liu G, Hong XY, Jiang ME, Yuan WW. Sustained-release G-CSF microspheres using a novel solid-in-oil-in-oil-in-water emulsion method. Int J Nanomed. 2012;7:4559–4569. doi: 10.2147/IJN.S33993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holman HYN, Martin MC, McKinney WR. Synchrotron-based FTIR spectromicroscopy: cytotoxicity and heating considerations. J Biol Phys. 2003;29:275–286. doi: 10.1023/A:1024465414395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang ZY, Chen M, Tong YJ, Ji T, Zhu HC, Peng WW. Performance of the infrared microspectroscopy station at SSRF. Infrared Phys Technol. 2014;67:521–525. [Google Scholar]
- 25.Mei Y, Miller L, Gao W, Gross RA. Imaging the distribution and secondary structure of immobilized enzymes using infrared microspectroscopy. Biomacromolecules. 2003;4:70–74. doi: 10.1021/bm025611t. [DOI] [PubMed] [Google Scholar]
- 26.Mohlenhoff B, Romeo M, Diem M, Wood BR. Mie-type scattering and non-Beer-Lambert absorption behavior of human cells in infrared microspectroscopy. Biophys J. 2005;88:3635–3640. doi: 10.1529/biophysj.104.057950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassan P, Byrne HJ, Bonnier F, Lee J, Dumas P, Gardner P. Resonant Mie scattering in infrared spectroscopy of biological materials—understanding the ‘dispersion artefact’. Analyst. 2009;134:1586–1593. doi: 10.1039/b904808a. [DOI] [PubMed] [Google Scholar]