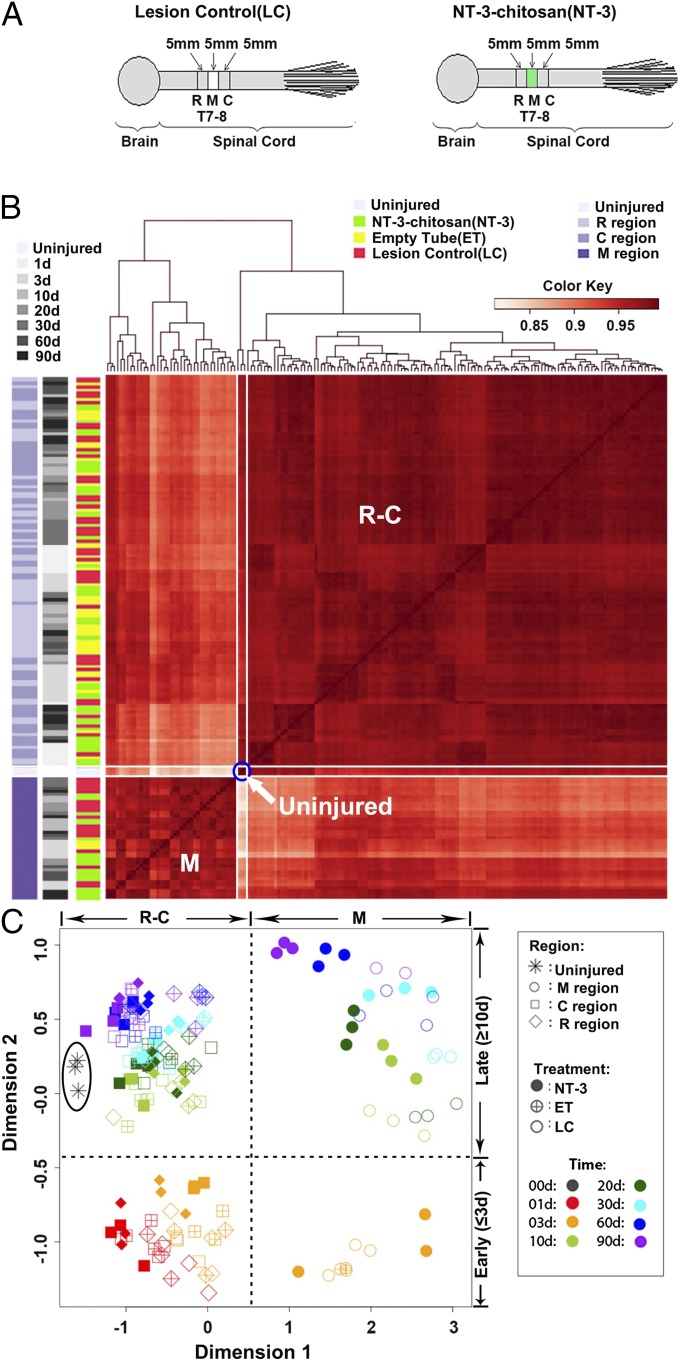
Fig. 1.
Sample clustering revealed temporal and spatial changes following SCI with or without NT3-chitosan treatment. (A) A schema describing anatomic positions of spinal cord segments for the LC and NT3 samples used for transcriptome analyses. Empty chitosan tubes (ET) not loaded with NT3 were included as well. Since no tissues were grown inside the empty chitosan tubes except at one early time point (i.e., day 3 postsurgery), ET samples were mostly present in R and C groups. (B) Heat map of hierarchical clustering of pairwise correlations among all 167 samples. The side color bars represent sample coding. Sample correlations among three uninjured samples are indicated by a blue circle. White-framed regions represent correlations between uninjured samples and all other injury samples in the M region (below the blue circle, with bottom-most samples in darker colors, i.e., better correlations) and R and C regions (above the blue circle, also with bottom-most samples showing better correlations). Note that the vertical and horizontal frames are symmetrical. Bottom-most and left-most M region samples were enriched for NT-3-chitosan treatment groups. (C) PCA of all samples.