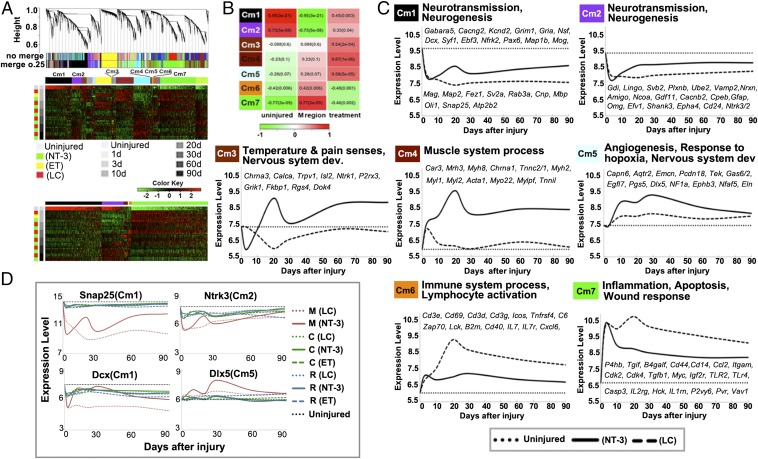
Fig. 3.
WGCNA of uninjured and all M region injury samples uncovered molecular mechanisms underlying the proregeneration function of NT3-chitosan. (A) Hierarchical cluster dendrogram using WGCNA analyzing uninjured samples (N) and samples in the M regions. A total of 19 modules were identified after 0.25 threshold merging. Heat maps underneath demonstrated gene expression of all 19 modules or 7 selected modules across uninjured and all M region injury samples. (B) Module trait correlation analysis revealed that seven modules were significantly correlated with NT3-chitosan treatment. (C) GO terms of each of the seven modules, averaged gene expression of top 30 most NT3-chitosan regulated genes within each module, and lesion controls across all time points in the M region were shown. Candidate genes within each module were listed. (D) Gene expression of four candidate genes belonging to Cm1, Cm2, and Cm5 across all 167 samples (all regions, all treatments, all time points). Note the robust effect of NT3-chitosan in M regions.