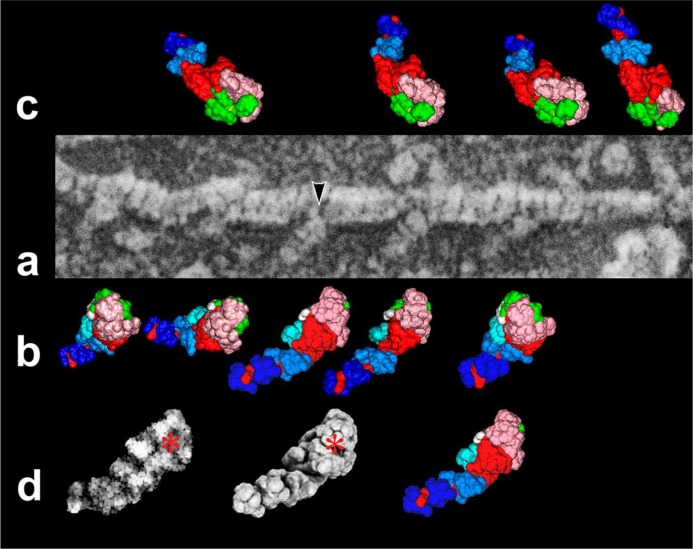
Figure 1.
a) Representative field of freeze-replicated actomyosin complex under rigor-state. All the actin-attached myosin heads show the configuration that matches rigor-form (1DFK), as defined by a comparison with simulated images. b)–c) Best-matched view for each particle along single actin filament is exhibited beside the original image. Particles on the bottom side (b) are overriding actin filament, whereas the very tip of those on the upper side (c) are partly hidden by actin. Subdomain composition of myosin S1 is color-coded as follows [upper-50 KDa, pink-tint; lower-50 K, green; N-terminal barrel, cyan; essential light-chain, sky-blue; regulatory light-chain, blue; 1st actin-contact site; yellow; 2nd actin-contact site, white; remaining part of heavy-chain including the lever-arm, red]. d) An example to examine the similarity of simulated image to replica image indicated by an arrowhead in (a). Left and center images represent the contrast-enhanced replica image and its best-matched simulation image generated by computer ray-tracing, respectively. The image texture had been pre-matched by morphological image-processing24. Note the good correlation between them. The asterisk indicates the position of the ATP-binding pocket. Subdomain constitution of the middle image was color-coded as above on the right. Here, the area of loop-2 (yellow) is much smaller than that in the actual molecule, due to the lack of atomic coordinates of that segment.