Figure 3. The ability of Mps1 to activate the SAC depends on its position in the kinetochore.
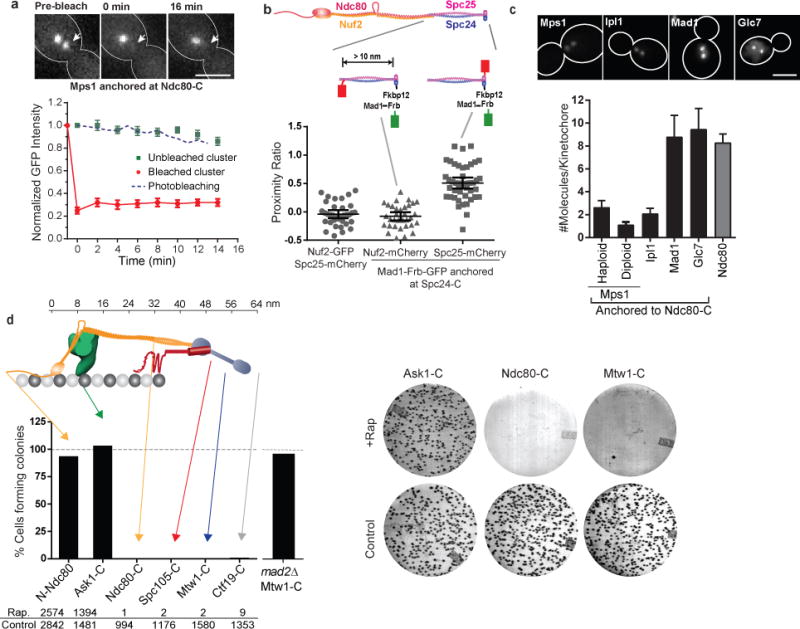
(a) Fluorescence recovery after photobleaching of Mps1-frb-GFP anchored at Ndc80-C (red circles), and loss of anchored protein from the unbleached cluster (green squares). Blue dashed line displays the expected rate of photobleaching as a result of imaging determined in cells expressing Ndc80-GFP (mean ± s.e.m. from n = 8 and 11 clusters for bleached and unbleached clusters, respectively; data pooled from 2 independent experiments). Scale bar ~ 3 μm.
(b) Top: Structure of Ndc80 complex and the positions of fluorescent tags used for FRET. Scatter plot: Proximity ratio, which is directly proportional to the FRET efficiency35, for FRET between Spc25-mCherry or Nuf2-mCherry and Mad1-Frb-GFP anchored to Spc24-C (mean ± 95% confidence interval from n = 35, 33 and 44 kinetochore clusters analyzed, from left to right. The experiment was repeated twice and graph presents mean data pooled from 2 independent experiments). Proximity ratio is defined as the acceptor fluorescence resulting from FRET normalized by the sum of direct excitation of mCherry on exciting GFP and GFP emission bleed-through into the mCherry imaging channel35. FRET between the anchored donor, Mad1-Frb-GFP, and the acceptor, Spc25-mCherry, was readily detected, but it was absent when the mCherry was fused to Nuf2-C. Spc25-C is < 3 nm away46 from Spc24-C, where the donor is anchored, whereas Nuf2-C is > 10 nm away63. We used Mad1, rather than Mps1, in this experiment to ensure that the number of donors is equal to the number of acceptor molecules (either Spc25-mCherry or Nuf2-mCherry, see (c) below) for accurate FRET quantification35.
(c) Number of protein molecules anchored at Ndc80-C, measured 30 min after rapamycin addition (mean ± s.d. from n = 25, 33, 29, 20, 41, and 30 kinetochore clusters from left to right. The experiment was performed once). Scale bar ~ 3 μm.
(d) Top: The organization of yeast kinetochore proteins along the microtubule axis20, 23. The N-terminal half of Spc105 is not drawn to scale. Bottom: Bar graph shows the number of colonies formed on rapamycin-containing plates relative to control plates. The experiment was repeated at least twice and the cumulative number of colonies scored is displayed below the graph. Right: Representative photographs of plates for three strains.
