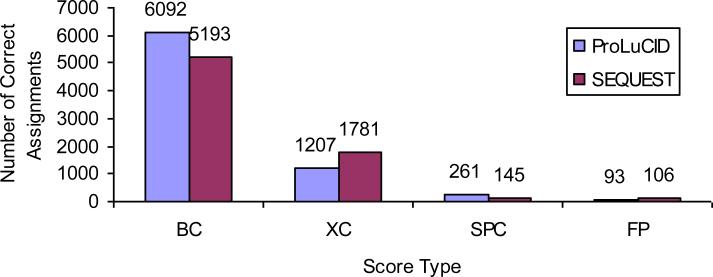
Figure 2.
Number of correct spectrum assignments by ProLuCID and SEQUEST XCorr and Sp scores. BC for both XCorr rank and Sp rank are correct; XC for XCorr rank is correct and Sp rank is incorrect; SPC for Sp rank is correct and XCorr rank is incorrect; FP for top hits on the reverse sequences of the 17 proteins. These results are based on a 6-step MudPIT with 75866 spectra. The ProLuCID XCorr outperforms SEQUEST XCorr in terms of number of correct spectrum assignments (7299 vs 6974); The ProLuCID Sp scores (binomial probability score) work better than SEQUEST Sp scores (6353 vs 5338); and ProLuCID XCorr gives more true hits the top rank than ProLuCID Sp (7299 vs 6353).