Figure 1. The pol β variant K167I is defective in polymerase activity.
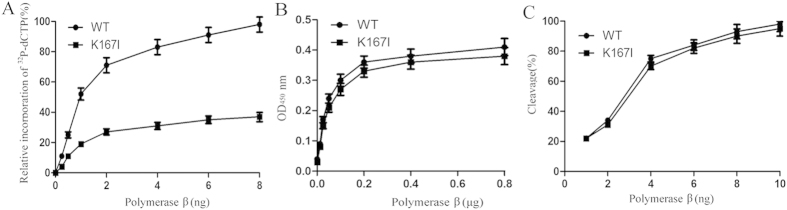
(A) Polymerase activity assays were performed with biotin labeled 1-nt gapped DNA substrate (pol-GAP). Separated DNA polymerization products were pulled down using Sepharose-avidin beads. After washing, the amount of radio labeled nucleotides incorporated into the products was determined by liquid scintillation counting. We found that the K167I variant (0.25 ng: 5.2 ± 0.57, 0.5 ng: 11.3 ± 1.25, 1 ng: 18.5 ± 2.31, 2 ng: 26.2 ± 2.94, 4 ng: 32.1 ± 3.51, 6 ng: 35.7 ± 3.97, 8 ng: 37.8 ± 4.84) had about 30% primer extension activity compared to the WT enzyme (0.25 ng: 11.2 ± 1.52, 0.5 ng: 25.4 ± 3.45, 1 ng: 52.6 ± 5.79, 2 ng: 71.8 ± 7.04, 4 ng: 84.5 ± 7.95, 6 ng: 91.3 ± 8.89, 8 ng: 99.1 ± 8.97). (B) ELISA based isotherm adsorption assays of the DNA bindi ng affinity of K167I (0.0125 μg: 0.08 ± 0.005, 0.025 μg: 0.15 ± 0.011, 0.05 μg: 0.21 ± 0.015, 0.1 μg: 0.27 ± 0.021, 0.2 μg: 0.33 ± 0.024, 0.4 μg: 0.36 ± 0.023, 0.8 μg: 0.38 ± 0.028) and WT pol β (0.0125 μg: 0.09 ± 0.005, 0.025 μg: 0.17 ± 0.010, 0.05 μg: 0.24 ± 0.015, 0.1 μg: 0.31 ± 0.021, 0.2 μg: 0.36 ± 0.020, 0.4 μg: 0.38 ± 0.023, 0.8 μg: 0.41 ± 0.028). The DNA substrate was biotin labeled pol-GAP. (C) Quantification of the 5′dRP lyase activity in K167I (1 ng: 22.4 ± 2.79, 2 ng: 31.7 ± 3.04, 4 ng: 70.6 ± 7.15, 6 ng: 82.1 ± 7.92, 8 ng: 89.7 ± 8.25, 10 ng: 95.4 ± 8.74) and WT pol β (1 ng: 21.9 ± 2.31, 2 ng: 35.2 ± 2.94, 4 ng: 76.2 ± 6.89, 6 ng: 84.3 ± 7.64, 8 ng: 93.1 ± 8.73, 10 ng: 98.7 ± 8.81) were shown.
