Fig. 1.
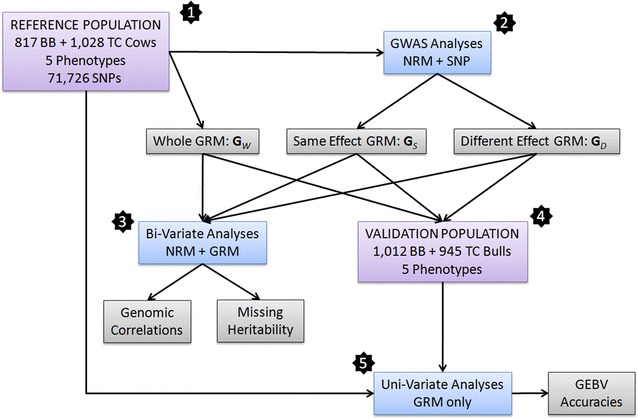
Flowchart of the analyses. 1 A reference population was created by merging data from 817 Brahman (BB) and 1028 Tropical Composite (TC) cows with measures on five traits and genotypes on 71,726 SNPs. A genomic relationship matrix (GRM) was constructed using the whole set of SNPs and termed G W. 2 GWAS analyses were performed separately for each breed using the pedigree-based numerator relationship matrix (NRM). Based on these GWAS, we compiled the set of SNPs with estimated effects in the same and opposite directions in the two breeds, the corresponding GRM were named G S and G D. 3 The three GRM were compared for their ability to produce estimates of genomic correlation and fractions of missing heritability using a series of bi-variate analyses, where the same phenotype was treated as a different trait in each breed and contained two additive genetic components, NRM and GRM. 4 A validation population was assembled by merging data from 1012 BB and 945 TC bulls with measures on the same five traits as for the cows in the calibration population. 5 The two populations, calibration and validation, were merged and after treating the phenotypes on the validation individuals as missing values, a series of uni-variate analyses (one for each trait) was undertaken with the GRM, either from G W, G S or G D as the only additive genetic component. Based on these analyses, the accuracies of the genomic predictions were computed and compared
