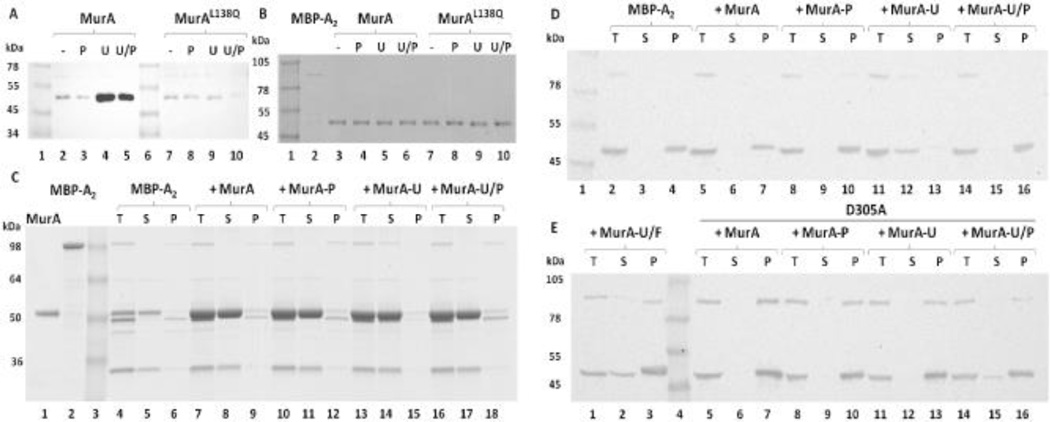
Figure 5. A2 binds MurA in a substrate-dependent manner.
(A) MBP-A2 associates with MurA in a closed conformational state. Eluates from amylose magnetic bead fractionation experiments were Western blotted and probed with the α-MurA antibody. MBP-A2 was incubated with MurA under various substrate conditions: No substrate (−), PEP (P), UDP-NAG (U), and both substrates (U/P). MurAL138Q rat mutant was tested in parallel. (B) Unbound fractions of panel A experiments. (C) Fusion cleavage analysis of A2-MurA binding. MBP-A2 was cleaved with TEV protease in the absence or presence of MurA under various substrate conditions (same as in panel A). Binding was assessed as A2 solubility after centrifugation: Total fraction (T), supernatant after centrifugation (S), and pellet fraction (P). Samples were resolved on SDS-PAGE. MBP-A2 has an apparent MW of 100 kDa. Cleaved A2 runs at 50 kDa with the MBP running at the same apparent MW as MurA (~52 kDa). (D) A2 binds MurA liganded to UDP-NAG. Western blot analysis of fusion cleavage assays. Blots were probed with the α-A2 antibody. Binding was tested with substrate conditions as in panel C. (E) A2 does not bind the tetrahedral intermediate state of MurA. UDP-NAG and Fosfomycin (U/F) liganded MurA and MurAD305A binding and immunoblotting was also performed in parallel as in panel D.