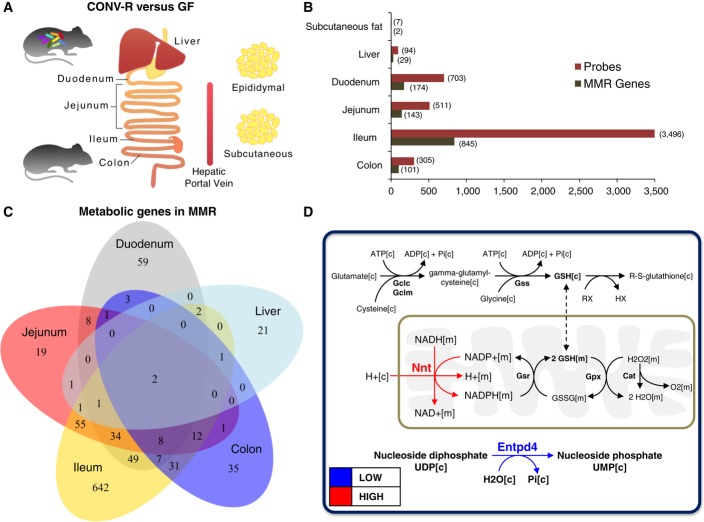
Figure 1.
- Liver as well as epididymal and subcutaneous WATs was obtained from both CONV-R and GF mice, and global gene expression profiling was generated using microarrays. Transcriptomics data for these three tissues were analyzed together with the previously published gene expression profiling of duodenum, jejunum, ileum, and colon tissues.
- Gene expression data for each tissue were normalized independently of other tissues, and significantly (Q-value < 0.05) differentially expressed probe sets and metabolic genes in Mouse Metabolic Reaction database were presented in each analyzed tissue.
- The overlap between the significantly (Q-value < 0.05) and differentially expressed metabolic genes in duodenum, jejunum, ileum, colon, and liver is presented.
- The significantly (Q-value < 0.05) and differentially expressed metabolic genes, Nnt, and Entpd4, as well as the reactions associated with Nnt, are presented. Red and blue arrows indicate the significantly higher (Q-value < 0.05) and lower expression of the metabolic genes in CONV-R mice compared to GF mice, respectively.