Figure 4.
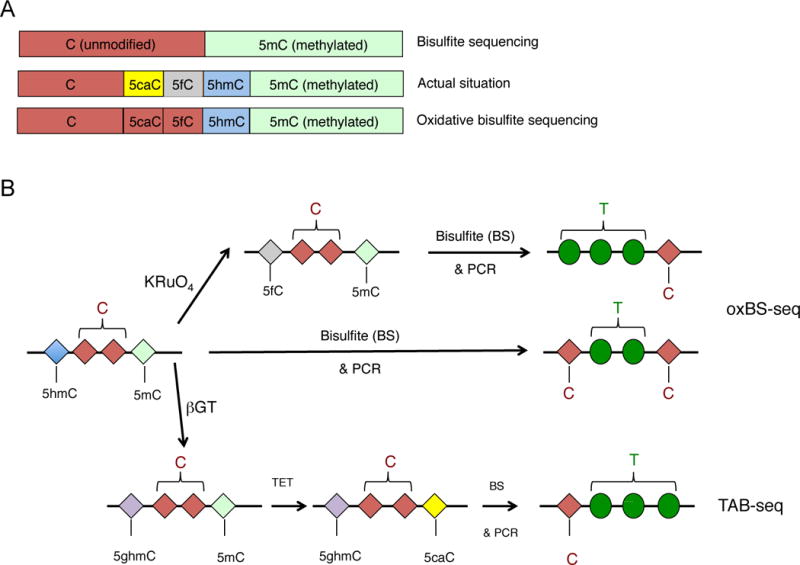
A. Top, Bisulfite sequencing (BS-seq) cannot discriminate methylated C from 5hmC, since both are converted after bisulfite treatment and PCR amplification to C. Middle, The actual state of C in the genome can be unmethylated C, 5caC, 5fC, 5hmC or 5mC. Bottom, The advent of oxidative bisulfite (oxBS) sequencing, which introduces chemical treatment of DNA prior to the bisulfite treatment, allows discrimination of 5hmC from 5mC, however the unmodified C is still not distinguished from 5cac or 5fC. B. Schematic representation of oxBS-seq and TAB-seq, two of the most useful methods that enable detection of 5hmC at single nucleotide resolution when combined with bisulfite sequencing. In the oxBS approach (top), DNA is treated with potassium perruthenate (KRuO4) that oxidizes 5hmC to 5fC. Subsequent bisulfite treatment converts 5fC (as well as 5caC and C) to U that will be read as T after PCR amplification, so only 5mC stays unconverted and is thus read as C after PCR amplification. In contrast in DNA samples that have been subjected exclusively to bisulfite treatment, both 5hmC and 5mC are read as C after PCR amplification, so 5mC, 5hmC and “unmodified” C can be discriminated by subtracting the results of BS-seq from those of oxBS. In TAB-seq (TET-assisted bisulfite sequencing), the DNA is treated with T2 phage beta-glucosyltransferase (βGT) that specifically recognizes and glycosylates 5hmC, generating glucosylated 5hmC (5ghmC). Subsequent treatment with purified and active TET1 leads to oxidization of 5mC to 5caC, and finally treatment of the sample with bisulfite results in complete conversion of all the bases to U, read as T after PCR amplification, with the exception of 5ghmC that stays unconverted and thus is read as C
