Abstract
Coordination between the Hertwig's epithelial root sheath (HERS) and apical papilla (AP) is crucial for proper tooth root development. The hedgehog (Hh) signaling pathway and Nfic are both involved in tooth root development; however, their relationship has yet to be elucidated. Here, we establish a timecourse of mouse molar root development by histological staining of sections, and we demonstrate that Hh signaling is active before and during root development in the AP and HERS using Gli1 reporter mice. The proper pattern of Hh signaling activity in the AP is crucial for the proliferation of dental mesenchymal cells, because either inhibition with Hh inhibitors or constitutive activation of Hh signaling activity in transgenic mice leads to decreased proliferation in the AP and shorter roots. Moreover, Hh activity is elevated in Nfic−/− mice, a root defect model, whereas RNA sequencing and in situ hybridization show that the Hh attenuator Hhip is downregulated. ChIP and RNAscope analyses suggest that Nfic binds to the promoter region of Hhip. Treatment of Nfic−/− mice with Hh inhibitor partially restores cell proliferation, AP growth and root development. Taken together, our results demonstrate that an Nfic-Hhip-Hh signaling pathway is crucial for apical papilla growth and proper root formation. This discovery provides insight into the molecular mechanisms regulating tooth root development.
KEY WORDS: Shh, Nfic, Proliferation, Root development, Mouse
Summary: During tooth development in mice, Nfic binds to Hhip to control Shh signaling, a process crucial for apical papilla growth and proper root formation.
INTRODUCTION
The tooth root is crucial for the physiological function of the dentition and oral health; it provides anchorage for the tooth and harbors sensory nerves in the pulp. Other important adjacent anatomical structures, such as the periodontal ligament (PDL) and alveolar bone, also develop together as a complex with the root. Understanding the processes of root development will help improve dental treatment via tooth regeneration approaches (Jussila and Thesleff, 2012). However, despite the importance of the root, its development has not been systematically investigated.
During development, the epithelial component of the tooth germ forms the Hertwig's epithelial root sheath (HERS), whereas the mesenchyme consists of the cranial neural crest cell (CNC)-derived dental papilla and dental follicle. In vivo tracing of CNC cells demonstrates that most of the root complex is formed by CNC-derived mesenchyme (Chai et al., 2000). The pulp and dentin are derived from the dental papilla, and the dental follicle gives rise to the cementum, PDL and adjacent alveolar bone. During root development, the dental papilla, together with the HERS, first grows as a soft tissue protrusion towards the apical side of the alveolar bone, followed by odontoblast differentiation to form the calcified dentin of the root (Bae et al., 2013a). The apical papilla (AP), the apical part of dental papilla, harbors mesenchymal stem cells (Sonoyama et al., 2008), and coordination between the HERS and AP is crucial for proper root development (Kumakami-Sakano et al., 2014). Previously, we have traced HERS cells during root development (Huang et al., 2009); however, to date there has not been an in vivo model for targeting the progenitor cell population from both the HERS and AP during root development.
Hh signaling is crucial for the development of many organs (Bai et al., 2004; Long et al., 2001; Treier et al., 2001). There are three Hh ligands in mammals: sonic hedgehog (Shh), Indian hedgehog (Ihh) and desert hedgehog (Dhh). In the quiescent condition when Hh ligand is absent, patched (Ptch), the Hh receptor, represses smoothened (Smo). When Hh ligand binds Ptch, it relieves the suppression of Smo. The accumulation of Smo in cilia then leads to the activation of Gli transcription factors and the downstream Hh signaling pathway (Briscoe and Thérond, 2013). This process is also fine-tuned by many regulators, one of which is hedgehog-interacting protein (Hhip). Hhip is a membrane protein that has similar binding affinity for Hh ligands as Ptch. By competing with Ptch for binding of Hh ligands, Hhip functions as an Hh attenuator (Chuang and McMahon, 1999). Previous studies have suggested that the Hh signaling pathway is involved in root development. Several molecules of the Hh signaling pathway are expressed during root development (Khan et al., 2007; Nakatomi et al., 2006). Moreover, mice with spontaneous Ptch1 mutation have short tooth roots and reduced proliferation around the HERS (Nakatomi et al., 2006).
Nuclear factor I/C (Nfic) is crucial for root development; Nfic−/− mice develop short molar roots with proliferation and odontoblast differentiation defects (Steele-Perkins et al., 2003). Nfic belongs to the Nuclear Factor-I (NF-I) family (Gronostajski, 2000), which regulates a large variety of genes. The four members of this family, Nfia, Nfib, Nfic and Nfix, share a common DNA binding sequence. The NF-I transcription factors bind to the dyad symmetric consensus sequence TTGGCNNNNNGCCAA and to individual half-sites with reduced affinity (Gronostajski et al., 1985; Meisterernst et al., 1988). Previous studies have found that NF-I transcription factors preferentially bind DNA target sites that are located close to transcription start sites and mainly act as activators of transcription of downstream target genes (Pjanic et al., 2011).
In this study, we provide in vivo evidence that Hh signaling participates in root development using lineage tracing and cell ablation of Gli1+ cells. We investigate Hh function in root development by both inhibition and constitutive activation of Hh activity. Finally, we demonstrate that Nfic promotes Hh attenuator Hhip, maintaining Hh activity in the apical papilla to sustain proper proliferation and apical growth of the root. Our study serves as an important contribution towards a better understanding of the regulatory mechanisms of tooth root development.
RESULTS
Postnatal root development and the identification of root progenitor cells
Although several reports have described root defect phenotypes following targeted gene mutation, there has not been a systematic analysis of postnatal root development (Bae et al., 2013b; Kim et al., 2013; Wang et al., 2013). Towards this end, we analyzed serial sagittal sections of the first mandibular molar in mice from PN0 to PN21 using HE staining. The typical section planes for the key stages are listed in supplementary material Fig. S1. We found that the crown continues to develop after birth until ∼PN4 (supplementary material Fig. S1A,B), at which point it reaches its full size. At PN7, the apical papilla begins to form a soft tissue protrusion towards the future alveolar socket (supplementary material Fig. S1C). The growth of soft tissue is followed by pre-odontoblast differentiation and dentin formation. Meanwhile, the PDL and alveolar bone under the root furcation and surrounding the root start to develop (supplementary material Fig. S1D). The root continues to elongate through PN14 (supplementary material Fig. S1E). By PN18, it has almost developed to its final length and the molar has erupted into the oral cavity (supplementary material Fig. S1F). Based on these findings, we used PN4 in our subsequent studies as the time point for the initiation of root development, and PN7 and 11 as time points for early stages of root development.
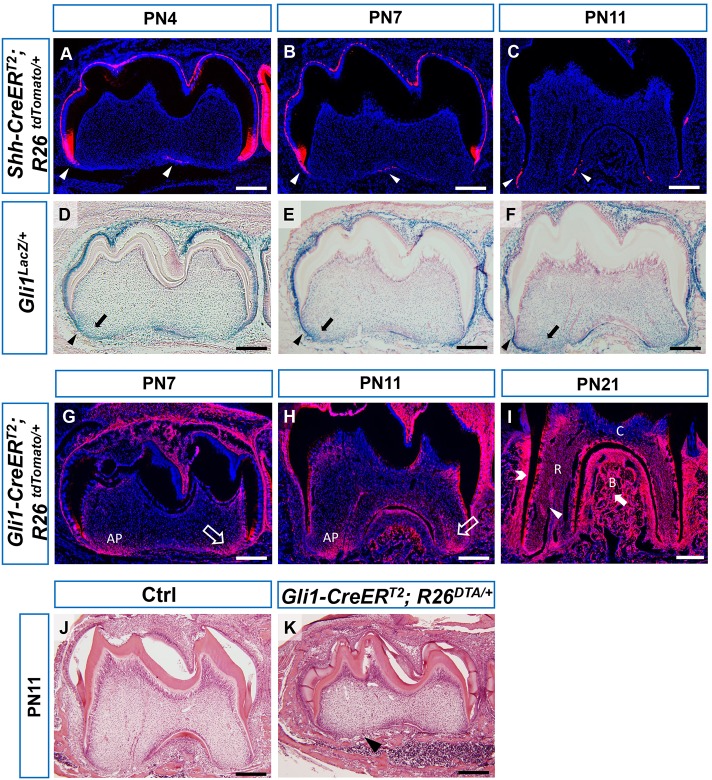
Next, we examined the expression pattern of Hh ligands during root development. Previous studies have reported that Shh is expressed in the dental epithelium at early stages, but it was difficult to detect Shh by immunostaining or in situ hybridization (ISH) at later stages, perhaps due to the length of the sample processing times (Li et al., 2015). To bypass this problem, we crossed Shh-CreERT2 with R26tdTomato/+ mice. Tamoxifen was injected ∼30 h before the collection times at PN4, 7 and 11. We optimized this timespan to balance two considerations: adequately expressing fluorescent protein for detection, and maintaining fidelity of reporter expression to reflect the original Shh-expressing cells rather than their descendants. The pattern of fluorescent reporter at early stages PN4 and PN7 was similar to what we reported for Shh ISH in our previous studies (Fig. 1A,B, arrowheads) (Li et al., 2015). We also detected Shh expression in the HERS at PN11 (Fig. 1C, arrowhead). Next, we examined Ihh expression using an IhhlacZ/+ reporter line. X-gal staining of IhhlacZ/+ mice revealed that Ihh was only secreted in bone at early stages (supplementary material Fig. S2, arrow), and, thus, was not likely to be involved in root development.
Fig. 1.
Gli1+ cells are root progenitor cells. (A-C) Sagittal sections of first mandibular molars of PN4, PN7 and PN11 Shh-CreERT2;R26tdTomato/+ reporter mice 30 h after tamoxifen injection. Arrowheads indicate fluorescent signal in the dental epithelium. (D-F) X-gal staining of first mandibular molars of PN4, PN7 and PN11 Gli1lacZ/+ mice. Arrowheads indicate strong signal in dental epithelium. Arrows indicate signal in apical papilla. (G-I) Lineage tracing of Gli1+ cells in first mandibular molars of PN7, PN11 and PN21 Gli1-CreERT2;R26tdTomato/+ mice after tamoxifen injection at PN3. Arrows indicate labeling of apical papilla in G-H. Arrowhead, open arrowhead and solid arrow indicate labeling of root pulp, PDL and alveolar bone, respectively. (J,K) HE staining of PN11 control (Ctrl) and Gli1-CreERT2;R26DTA/+ mice treated with tamoxifen at PN3. Arrowhead indicates lack of root formation. n=3 per group. AP, apical papilla; C, crown; R, root; B, alveolar bone. Scale bars: 200 µm.
After confirming the presence of Hh ligand, we analyzed Gli1 expression using a Gli1lacZ/+ reporter line. Gli1 is a direct transcription target of Hh, so it serves as readout of Hh activity levels. We found that the Gli1 expression pattern and level after X-gal staining of Gli1lacZ/+ mice were consistent with the Shh expression pattern and previous studies of Gli1 ISH (Khan et al., 2007). The area of highest Gli1 expression co-localized with Shh+ cells in the epithelial component of the tooth germ (Fig. 1D-F, arrowheads). Gli1 expression was also detectable in mesenchymal cells close to the region of Shh expression (Fig. 1D-F, arrows), and Gli1 intensity in the pulp appeared to diminish as the distance from the source of Shh increased.
These expression patterns provided the basis for our lineage tracing approach, because (1) root progenitor cells reside in the HERS, apical papilla and dental follicle, and (2) these regions are Gli1+ at PN4, the initiation stage. We hypothesized that we would be able to target the root progenitor cells using a Gli1-CreERT2/loxP system. We crossed Gli1-CreERT2 mice with R26tdTomato/+ fluorescent reporter mice to create Gli1-CreERT2;R26tdTomato/+ mice. After inducing the mice with tamoxifen just before root initiation at PN3, we found that the root complex, including the root pulp, PDL and surrounding alveolar bone, was positive for the fluorescent reporter during (Fig. 1G,H) and after (Fig. 1I) root development. To test further whether Gli1+ cells give rise to the root, we performed cell ablation assays. Gli1-CreERT2;R26DTA/+ mice were injected with tamoxifen at PN3 to ablate Gli1+ cells before root development. These pups failed to survive to adulthood, but survived long enough to determine that their tooth roots had failed to form by PN11 (Fig. 1K, arrowhead), whereas roots of control mice had already begun to develop (Fig. 1J). Immunostaining of caspase-3 showed elevated apoptosis in the tooth germ of Gli1-CreERT2;R26DTA/+ mice (supplementary material Fig. S3B, arrowhead). Thus, based on the lineage tracing and cell ablation, we conclude that Gli1+ cells appear to be root progenitor cells and are crucial for root development. The Gli1-CreERT2/loxP system can target these root progenitor cells during root development.
Proper levels of Hh signaling activity are crucial for apical papilla growth
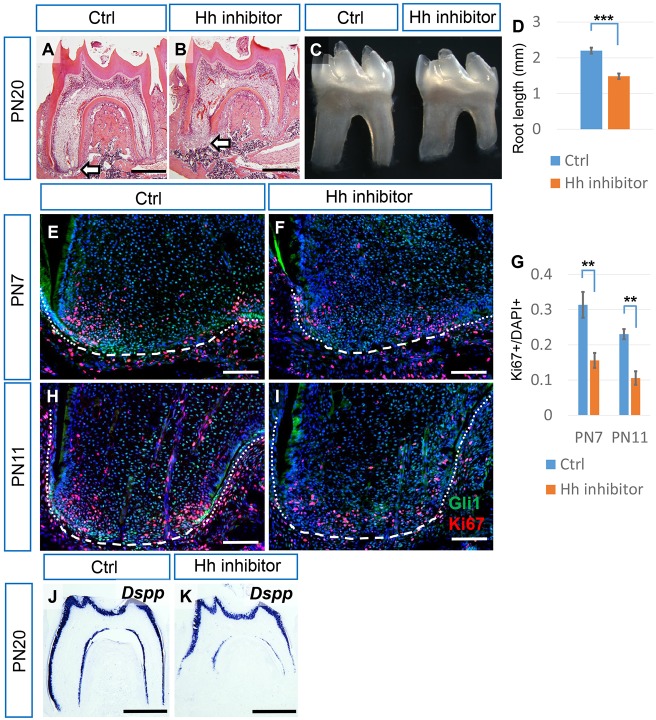
In order to investigate the function of Hh signaling during root development, we manipulated Hh activity both negatively and positively. First, we inhibited Hh by injecting small-molecule Hh inhibitors (Zhao et al., 2015). After treating mice with an Hh inhibitor, the root complex exhibited relatively normal morphology (Fig. 2A-C); however, root length was reduced at PN20 from 2.20±0.081 mm (sum of mesial and distal root length, n=3 per group) untreated to 1.48±0.075 mm with treatment (Fig. 2D). At PN7, Hh activity was repressed after Hh inhibitor treatment based on Gli1 activity (Fig. 2E,F), and proliferation was downregulated in the apical region (Fig. 2E-G). At the later developmental stage of PN11, Hh activity and proliferation were also reduced in molars treated with inhibitor (Fig. 2G-I). Moreover, the odontoblast differentiation marker Dspp was downregulated in roots after treatment with Hh inhibitor (Fig. 2J,K). We also analyzed apoptosis by analyzing caspase-3 activity, but apoptosis was undetectable in the apical papilla of molars treated or untreated with Hh inhibitor (unpublished data).
Fig. 2.
Inhibition of Hh signaling results in reduced proliferation and odontoblast differentiation and shorter roots. (A,B) HE staining of the first mandibular molar from PN20 control mice untreated (Ctrl) or treated with Hh inhibitor. Arrows indicate the apical end of the mesial root. (C) Macroscopic view of dissected first mandibular molars from PN20 control mice untreated (Ctrl) or treated with Hh inhibitor. (D) Quantification of the sum of the mesial and distal root lengths of first mandibular molars at PN20 from micro-CT. n=3 per group. (E,F,H,I) Immunostaining of Gli1 (green) and Ki67 (red) in the mesial apical papilla of first mandibular molars from PN7 and PN11 control mice untreated (Ctrl) or treated with Hh inhibitor. Dotted lines indicate the HERS, dashed lines indicate the border of the apical papilla. (G) Quantification of Ki67+ nuclei/total nuclei from E,F and H,I. n=3 per group. (J,K) In situ hybridization of Dspp in the first mandibular molars of PN20 control mice untreated (Ctrl) or treated with Hh inhibitor. Scale bars: 500 µm in A,B,J,K; 100 µm in E,F,H,I. **P<0.01; ***P<0.001.
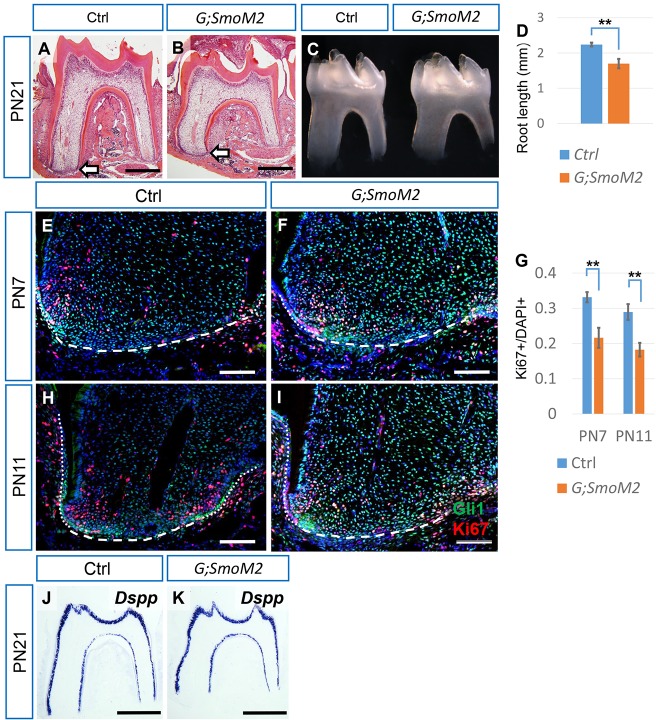
Next, we investigated the effect of elevated Hh activity during root development. We used Gli1-creERT2;R26SmoM2fl/fl mice induced with tamoxifen at PN3 to produce constitutive expression of Smo, resulting in unrestrained Hh activity in root progenitor cells. Surprisingly, overactivity of Hh recapitulated the phenotype of inhibition of Hh. Although the root and surrounding structures developed relatively normally after constitutive activation of Hh (Fig. 3A-C), root length was reduced (1.70±0.134 mm versus 2.24±0.056 mm, n=3 per group) (Fig. 3D). Again, we analyzed Gli1 and Ki67 as indicators of Hh activity and proliferation status, respectively. At both PN7 and 11, Gli1 expression in the apical papilla was upregulated (Fig. 3E,F,H,I) and proliferation was reduced (Fig. 3E-I). Odontoblast differentiation marker Dspp expression was unaffected by constitutive activation of Hh (Fig. 3J,K). We also analyzed apoptosis by analyzing caspase-3 activity, but apoptosis was undetectable in the apical papilla of molars in both groups (unpublished data). Taken together, we found that both inhibition and constitutive activation of Hh signaling in the apical papilla affected proliferation and resulted in reduced root length.
Fig. 3.
Constitutive activation of Hh signaling in root progenitor cells leads to reduced proliferation and shorter roots. (A-C) HE staining and macroscopic views of first mandibular molars from PN21 control (Ctrl) and Gli1-CreERT2;R26SmoM2fl/fl (G;SmoM2) mice after tamoxifen injection at PN3. Arrows indicate the apical end of the mesial root. (D) Quantification of the sum of the mesial and distal root lengths of first mandibular molars at PN21 from micro-CT. n=3 per group. (E,F,H,I) Immunostaining of Gli1 (green) and Ki67 (red) in the mesial apical papilla of first mandibular molars from PN7 and PN11 control and G;SmoM2 mice. Dotted lines indicate the HERS, dashed lines indicate the border of the apical papilla. (G) Quantification of Ki67+ nuclei/total nuclei from E,F and H,I. n=3 per group. (J,K) In situ hybridization of Dspp in the first mandibular molars of PN21 control and G;SmoM2 mice. Scale bars: 500 µm in A,B,J,K; 100 µm in E,F,H,I. **P<0.01.
Nfic inhibits the Hh signaling pathway via Hhip
Previous studies have shown that Nfic−/− mice are an in vivo model for root development defects, so we tested whether Hh is involved in the growth defect of roots in Nfic−/− mice. We examined Hh activity in the root by analyzing Gli1 activity and found that Gli1 expression was elevated in the apical papilla of Nfic−/− mice at both PN7 (Fig. 4A,B) and PN11 (Fig. 4C,D), reminiscent of the phenotype in the apical papilla after constitutive activation of Hh. We confirmed the elevation of Hh activity using qRT-PCR (Fig. 4E). By contrast, we also examined Nfic expression after treatment with Hh inhibitor and found no significant change, based on in situ hybridization and qPCR (supplementary material Fig. S4).
Fig. 4.
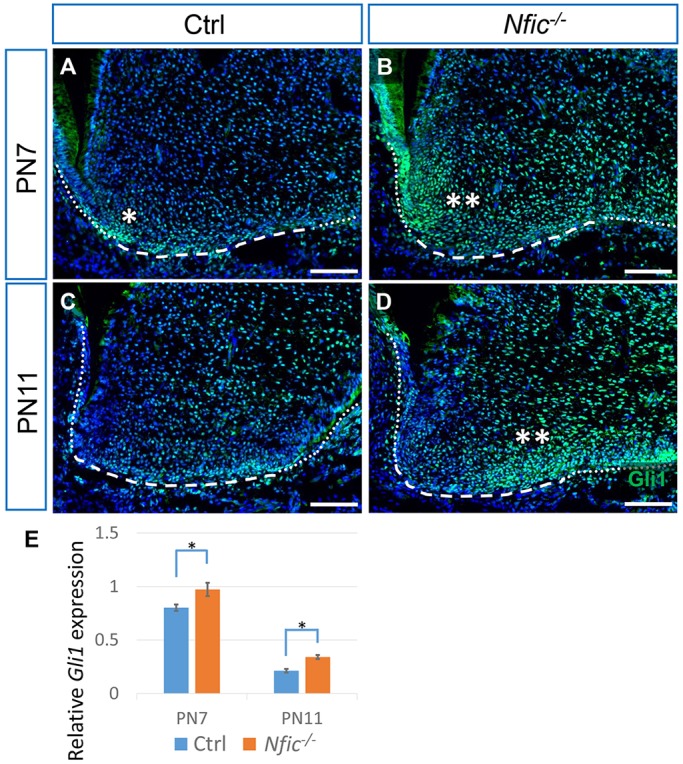
Hh activity is elevated in the apical papilla of Nfic–/– mice. (A-D) Immunostaining of Gli1 (green) in the mesial apical papilla of first mandibular molars from PN7 and PN11 control (Ctrl) and Nfic–/– mice. Dotted lines indicate the HERS, dashed lines indicate the border of the apical papilla. Single asterisk indicates positive staining, double asterisk indicate elevated staining signal. (E) Quantitative RT-PCR of Gli1 from the apical tissue of first mandibular molars of control and Nfic–/– mice at PN7 and PN11. n=3 per group. Scale bars: 100 µm. *P<0.05.
To investigate how Nfic regulates Hh, we first tested which genes were altered in the molar tooth germs of Nfic−/− mice at PN4 using RNA-sequencing (GEO accession GSE69054). We identified a list of 275 genes as differentially expressed in Nfic−/− mice (40 genes upregulated, 235 genes downregulated) (supplementary material Table S2). In that list, we searched for genes related to the Hh signaling pathway, including ligands, receptors, transcriptional effectors, regulators and other mediators. The Hh negative regulator Hhip was significantly reduced in Nfic−/− mice (P=0.00258). Next, we validated this candidate target gene by examining its expression pattern using RNAscope in situ hybridization analysis. At early stage of root initiation, Shh was expressed in the dental epithelium, whereas Gli1 and Hhip were expressed in both the dental epithelium and mesenchyme (Fig. 5A-D). Despite background fluorescence in the dental epithelium, expression of Nfic and Hhip was detectable in the dental papilla, consistent with a possible interaction between these two genes (Fig. 5E). Moreover, Hhip expression was downregulated in the dental papilla of Nfic−/− mice (Fig. 5F,G). Previous studies have demonstrated that NF-I transcription factors preferentially bind their DNA target sites around transcription start sites and that they mainly act as activators of transcription (Pjanic et al., 2011). Therefore, we searched for NF-I binding sites within 2.5 kb upstream of the transcription start site of Hhip using two different web servers. Twenty-two putative binding sites for NF-I were predicted from PROMO (http://alggen.lsi.upc.es), and seven putative binding sites for NF-I were predicted by Matinspector (www.genomatix.de), with two sites in common (NF-I binding site 1: −1456 to −1452; site 2: −527 to −524). After designing primers for these two sites, we performed ChIP assays (Fig. 5H). For both binding sites, chromatin fragments were immunoprecipitated with anti-NF-I antibody but not with control antibody. Taken together, our results suggest that Nfic inhibits Hh activity by promoting the Hh attenuator Hhip.
Fig. 5.
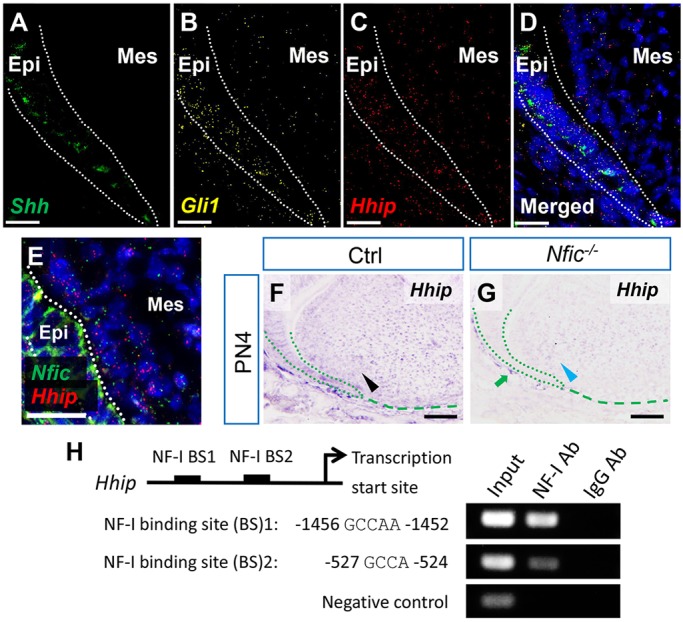
Nfic regulates the Hh signaling pathway via Hhip. (A-D) RNAscope in situ hybridization (ISH) of Shh, Gli1 and Hhip in first mandibular molars from PN4 control mice. Dotted lines indicate the dental epithelium. (E) RNAscope ISH of Nfic and Hhip in first mandibular molars from PN4 control mice. Dotted lines indicate the dental epithelium. (F,G) ISH of Hhip in first mandibular molars from PN4 control (Ctrl) and Nfic–/– mice. Black arrowhead indicates positive signal in the dental papilla. Blue arrowhead indicates reduced signal in the dental papilla. Green arrow indicates positive signal in the dental follicle. Green dotted lines indicate the dental epithelium, green dashed lines indicate the apical border of the dental papilla. (H) Schematic diagram of the upstream region of the mouse Hhip gene shows locations of putative NF-I binding sites tested in ChIP assays (not to scale). Putative binding sequences are shown below. (Right) ChIP assays of chromatin fragments immunoprecipitated with anti-NF-I antibody (Ab) or IgG control antibody. Immunoprecipitates were PCR-amplified, with primers flanking the putative binding sites or negative control region without binding site. Input lane shows PCR amplification of chromatin fragments without immunoprecipitation. n=3 per group. Epi, epithelium; Mes, mesenchyme. Scale bars: 25 µm in A-E; 100 µm in F,G.
Attenuation of elevated Hh activity partially rescues proliferation and root development in Nfic−/− mice
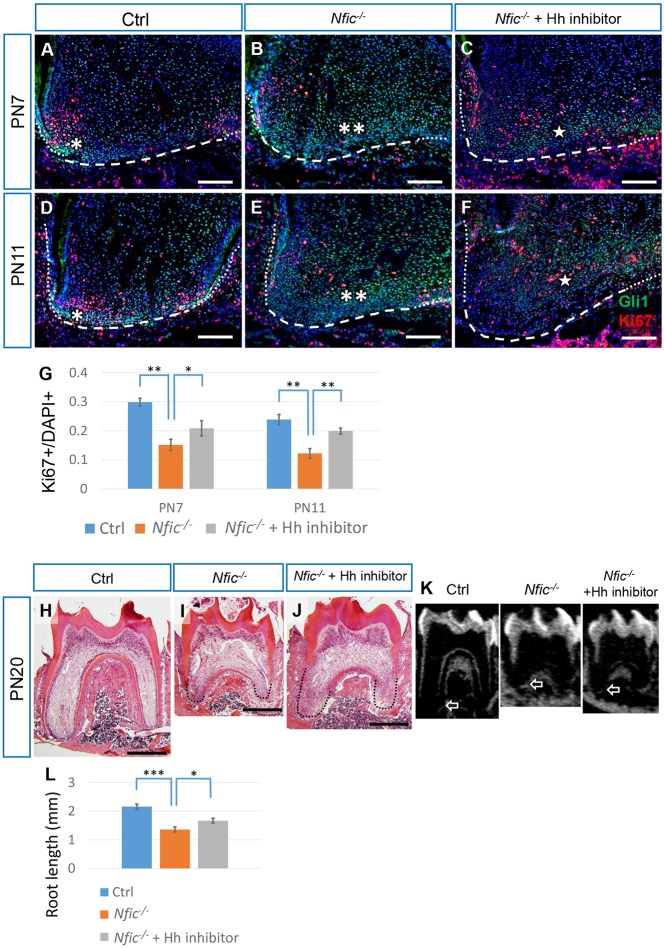
We hypothesized that the elevated Hh activity in Nfic−/− mice resulted in a growth defect of the apical papilla. To test this hypothesis, we applied Hh inhibitor to Nfic−/− mice and evaluated whether reversing the elevated Hh activity in the apical papilla would rescue the proliferation defect in the apical papilla and the short root length. As expected, treatment with Hh inhibitor resulted in an attenuation of Gli1 expression at PN7 (Fig. 6B,C) and PN11 (Fig. 6E,F). Moreover, the proliferation defect in the apical papilla of Nfic−/− mice was also partially rescued at PN7 (Fig. 6B,C,G) and PN11 (Fig. 6E-G) following Hh inhibitor treatment. Although the dentin defect was not rescued (Fig. 6H-J), Nfic−/− pups treated with Hh inhibitor developed longer roots than untreated pups did (1.70±0.053 mm versus 1.37±0.101 mm; n=9 for Nfic−/− mice treated with Hh inhibitor, n=5 for Nfic−/− mice; control 2.15±0.093 mm, n=5) (Fig. 6K,L), indicating that the growth defect in the roots of Nfic−/− mice was partially rescued by reversing the elevated Hh activity.
Fig. 6.
Attenuation of elevated Hh activity partially rescues the cell proliferation and root defects in Nfic–/– mice. (A-F) Immunostaining of Gli1 (green) and Ki67 (red) in the mesial apical papilla of first mandibular molars from PN7 and PN11 control mice (Ctrl), Nfic–/– mice and Nfic–/– mice treated with Hh inhibitor. Dotted lines indicate the HERS, dashed lines indicate the border of the apical papilla. Single asterisk indicates Gli1 signal; double asterisks indicate elevated Gli1 signal; single five-point star indicates attenuated Gli1 signal. (G) Quantification of Ki67+ nuclei/total nuclei from A-F. n=3 per group. (H-K) HE staining and micro-CT slice views of first mandibular molars from PN20 control mice (Ctrl), Nfic–/– mice and Nfic–/– mice treated with Hh inhibitor. Dotted line indicates the root in H-J. White arrows indicate the apical end of the mesial root in K. (L) Quantification of the sum of the mesial and distal root lengths of first mandibular molars at PN20 from micro-CT. n=5 for Ctrl and Nfic–/– mice, n=9 for Nfic–/– mice treated with Hh inhibitor. Scale bars: 100 µm in A-F; 500 µm in H-J. *P<0.05; **P<0.01;***P<0.001.
DISCUSSION
This study expands our understanding of the relationship between the HERS and apical papilla during root development. We have found that epithelial cells in the HERS secrete Shh ligand and that Nfic activates Hh attenuator Hhip in the mesenchyme, creating an Hh activity gradient that results in proper proliferation and growth patterns of the apical papilla during root development. Our results explain why roots in Nfic−/− mice form a normal HERS structure (Park et al., 2007) and Shh ligand expression is not significantly altered (our RNA sequencing data), but they exhibit a root defect because mesenchymal cells fail to respond correctly to the Shh signal from the epithelium without Nfic-activated Hhip to regulate this process. This model also explains our previous finding that adding Shh beads fails to rescue the root defect observed in the Nfic−/− tooth germ (Huang et al., 2010). Without Nfic-activated Hhip, Hh activity is unrestrained in the mesenchyme, and adding additional Shh only exacerbates the situation. Moreover, there might be a negative-feedback loop in which Nfic as an Hh inhibitor is indirectly induced by Shh. This putative feedback loop might explain why we detected Nfic expression around Shh beads in the dental pulp of tooth germs (Huang et al., 2010).
To date, reports of root defects in mice have mostly been based on approaches using a non-inducible cre/loxP system, which is not tailored for studying root development. Some models, such as Osr2-Cre;Smad4fl/fl (Li et al., 2011) and K14-Cre;Smad4fl/fl (Huang et al., 2010), show phenotypes in early stages of tooth development, but it is hard to determine whether the root defect is secondary to the crown defect. Other models, including OC-Cre (Bae et al., 2013b; Gao et al., 2009; Kim et al., 2013; Zhang et al., 2013), Col1a1-Cre (Kim et al., 2012) and Sp7-Cre (Rakian et al., 2013; Wang et al., 2013), take effect too late, only targeting differentiated cells. Here, we used the Gli1-CreERT/loxP system to target the early progenitor cells for root development. We are confident that this model will facilitate future studies on root development.
We used Shh-CreERT2;R26tdTomato/+ mice induced with tamoxifen 30 h prior to harvesting samples in order to evaluate Shh expression. Although we have successfully detected Shh with immunostaining in nerves of adult mice (Zhao et al., 2014), it was difficult to obtain a clear Shh signal with immunostaining or ISH in the developing molar root (Li et al., 2015), probably due to the limited amount of HERS tissue and the time required for sample processing. The Shh expression pattern in Shh-CreERT2;R26tdTomato/+ mice at early stages was consistent with our previous Shh ISH results (Li et al., 2015).
Hh signaling pathway has been reported to regulate tooth development at early stages. Gli2/Gli3 double knockout mice failed to form molar tooth germ (Hardcastle et al., 1998). Conditional knockout of Shh or Smo in dental epithelium both caused fused and abnormal molar tooth germs (Dassule et al., 2000; Gritli-Linde et al., 2002). Evc and Ift88, genes related to cilia and Hh signal transduction, also regulated normal patterning of molar tooth germ (Nakatomi et al., 2013; Ohazama et al., 2009). Our study suggests that Hh also plays a key role in later stages of tooth development.
Previous studies have proposed that Shh acts as a graded morphogen, such as in neural tube differentiation (Dessaud et al., 2008; Ribes and Briscoe, 2009) and digit patterning (Harfe et al., 2004). Different dosages of Shh trigger different reactions in recipient cells (Harfe et al., 2004). Shh has a dual effect on retinal ganglion cell axonal growth, acting as a positive factor at low concentrations and a negative factor at high concentrations (Kolpak et al., 2005). Moreover, Hh stimulates proliferation via a paracrine or juxtacrine mechanism, because high levels of proliferation were detectable in cells adjacent or in close proximity to Smo-expressing cells in the mammary gland (Visbal et al., 2011). Similarly, Gli1 appears to be expressed in a gradient in the HERS and apical papilla of molar roots, with the strongest Gli1 expression in the HERS. In the adjacent mesenchymal cells, Gli1 intensity appears to diminish gradually as the distance increases. Likewise, the most active proliferation in the apical papilla takes place in the mesenchymal cells near the HERS, in which there is a switch from high Gli1 intensity to low Gli1 intensity away from the HERS. We believe that a gradient model for Hh activity and cell proliferation might explain our results using an Hh inhibitor or ectopically activating Hh in mesenchyme cells of the apical papilla. Either treatment dampens the Hh gradient, resulting in a cell proliferation defect. By contrast, loss of Smad4 in the dental epithelium results in increased Shh expression in the epithelium during root development and upregulated cell proliferation (Li et al., 2015). We propose that increased Shh in the epithelium results in a greater differential gradient of Hh activity level between the epithelium and nearby mesenchymal cells, and therefore increased cell proliferation.
Hh has also been reported to play a role in differentiation. Gli-CreERT2;Smofl/fl mice exhibit osteoporosis and a reduction in bone volume in craniofacial bones and odontoblast defects in incisors (Zhao et al., 2015). In our study, Hh inhibitor reduced the expression level of odontoblast differentiation marker Dspp in molar roots, which might be due to a function of Hh in odontoblast differentiation, or might be partially secondary to the apical papilla growth defect. However, OC-Cre;R26SmoM2fl/fl mice also had impaired development of postnatal bone, which became apparent 3 weeks after birth (Cho et al., 2012). Similarly, in early stage of craniofacial development, both knockout Smo (Wnt1-Cre;Smon/c) and constitutive activation of Hh (Wnt1-Cre;R26SmoM2) in neural crest cells disrupted normal head skeleton (Jeong et al., 2004). All these results might represent additional examples of a requirement for the proper balance of Hh activity, in this case being crucial for patterning and differentiation. In our current study, because root development completed within 2 weeks, we did not detect a significant odontoblast differentiation defect in Gli1-creERT2;R26SmoM2fl/fl mice, suggesting that the reduced root length was not likely to be secondary to osteoblast or odontoblast differentiation defects.
We have demonstrated that an Nfic-Hhip-Hh signaling cascade controls proliferation in the apical papilla. The NF-I-Hhip-Hh signaling cascade appears to play a role in other organs as well, because Nfib−/− mice and Hhip−/− mice show similar defects in lung development (Chuang et al., 2003; Steele-Perkins et al., 2005). Multiple signaling pathways have been implicated in regulating root development (Lohi et al., 2010), and Nfic clearly regulates root development through multiple target genes; thus, more pathways remain to be uncovered. Previous studies have shown that Nfic−/− mice also display a root dentin defect (Lee et al., 2009) and have identified candidate genes as targets of Nfic in odontoblast differentiation (Lee et al., 2014; Zhang et al., 2015a).
In summary, we have identified the involvement of the Nfic-Hhip-Hh cascade in root development and partially rescued the growth defect in Nfic−/− mice by reducing Hh activity. This study further expands our understanding of Hh function in development and reveals its association with proper proliferation and growth. Our findings might offer potential instruction for tissue engineering and stem cell-mediated tooth regeneration in the future.
MATERIALS AND METHODS
Animals
All animal information (sources and original references) is listed in supplementary material Table S1. All mouse experiments were conducted in accordance with protocols approved by the Department of Animal Resources and the Institutional Animal Care and Use Committee of the University of Southern California, USA.
Tamoxifen and Hh inhibitor administration
Tamoxifen (Sigma, T5648) was dissolved in corn oil (Sigma, C8267) and injected intraperitoneally at post-natal day (PN) 3 for Gli1-CreERT2;R26tdTomato/+, Gli1-CreERT2;R26DTA/+ and Gli1-CreERT;R26SmoM2fl/fl mice or 30 h prior to indicated harvest time for Shh-CreERT2; R26tdTomato/+ mice with a single injection at 200 mg/kg body weight.
Hh inhibitor GDC-0449 (Selleckchem, S1082) was dissolved in dimethyl sulfoxide (DMSO, Sigma, D2650) and injected intraperitoneally once a day at 24 mg/kg body weight from PN3 to PN13 for the Hh inhibitor assay, and from PN3 to PN9 for Nfic−/− mice. Control pups were injected with the same dosage of DMSO alone.
X-gal staining
Mandibles (n=3 per stage) were dissected under a stereomicroscope (Leica L2), fixed in 0.2% glutaraldehyde in PBS with 2 mM MgCl2 overnight at 4°C, and decalcified in 10% EDTA in PBS with 2 mM MgCl2 for 7 days at 4°C. After dehydration in 30% and then 60% sucrose solutions at 4°C until the sample sank to the bottom, the samples were soaked in OCT compound (Sakura Tissue-Tek, 4583) at 4°C for 6 h before embedding. Cryosectioning was performed to obtain slices of 8 μm thickness using a cryostat (Leica CM1850), followed by X-gal staining according to standard protocol, as previously described (Li et al., 2015).
Fluorescence and histological analysis
Samples (n=3 per stage) were dissected and fixed in 4% paraformaldehyde in PBS overnight at 4°C, then decalcified in 10% EDTA in PBS for 7 days at 4°C in the dark. For fluorescent reporter observation, samples were dehydrated, embedded in OCT and sectioned as described above for X-gal staining. Sections were counterstained with DAPI solution and mounted with fluoro-gel (Electron Microscopy Sciences, 17985-10). Images were captured using a fluorescence microscope (Leica DMI 3000B).
For double immunostaining, fluorescent reporter samples were processed as described above. The sections were first incubated with blocking reagent (PerkinElmer, FP1012) for 2 h at room temperature, then incubated with anti-Gli1 antibody (Novus Biological, NBP1-78259; 1:500) at 4°C overnight, followed by HRP-labeled goat anti-rabbit IgG (PerkinElmer, NEF812001EA; 1:200) and TSA kits (PerkinElmer, NEL741001KT). Next, immunostaining for Ki67 was performed with anti-Ki67 antibody (Abcam, ab16667; 1:50) and Alexa Fluor 568 goat anti-rabbit IgG (Invitrogen, A11011; 1:200). Immunostaining for Caspase-3 was performed with anti-active and pro-Caspase 3 antibody (Abcam, ab13847; 1:150). Sections were counter-stained, mounted and imaged as described above. Positively stained nuclei in the apical papilla region were counted using Image-Pro Plus 6.0 software.
For HE staining, samples were fixed, decalcified, dehydrated with serial ethanol and embedded in paraffin. Sections were cut to a thickness of 5 μm and HE staining was performed according to standard procedures.
RNAscope in situ hybridization (ISH)
For RNAscope ISH, PN4 samples (n=3) were dissected and fixed in fresh 10% neutral buffered formalin at room temperature for 24 h, then dehydrated with serial ethanol and embedded in paraffin. Sections were cut to a thickness of 5 μm, and staining was performed with RNAscope Multiplex Fluorescent kit (Advanced Cell Diagnostics, 320850) according to the manufacturer's instructions. Shh, Gli1, Hhip and Nfic probes for RNAscope Multiplex Fluorescent assay were designed and synthesized by Advanced Cell Diagnostics.
To create the Hhip probe, a plasmid containing mouse full-length Hhip cDNA was kindly provided by Andrew McMahon (University of Southern California). Primers were designed to amplify a 700 bp fragment (NCBI reference, NM_020259.4, nt 108-810) for the probe template. An SP6 sequence was added at the start of the forward primer and a T7 sequence was added to the start of the reverse primer. The primer sequences were: SP6-forward, 5′-GCTATTTAGGTGACACTATAGAAGTGcctttcggttcctgctactg-3′; T7-reverse, 5′-TTGTAATACGACTCACTATAGGGAGAagcctgctgcattctgagtt-3′. The PCR product was extracted and purified (Qiagen, 28704) from a 1.5% agarose gel after electrophoresis. RNase Inhibitor (Roche, 03335399001), DIG RNA Labeling Mix (Roche, 11277073910) and T7 RNA polymerase (Roche, 10881767001) were used to create the antisense RNA probe. Nfic and Dspp probes were synthesized as previously described (Huang et al., 2010). Samples (n=3 per stage) were fixed, decalcified, dehydrated, embedded and sectioned as described above for HE staining. ISH was performed with standard protocols, as previously described (Li et al., 2015).
Micro-CT analysis
Mandibles were dissected and fixed in 4% paraformaldehyde in PBS overnight at 4°C before micro-CT analysis. Micro-CT scans and X-ray pictures were taken by SkyScan 1174 micro-CT (Bruker) at a resolution of 9.6 μm. Images were reconstructed using AVIZO 7.1 (Visualization Sciences Group), and root lengths were measured along the mesial border of mesial root and the distal border of distal root from the end of enamel to the bottom of alveolar socket.
Quantitative reverse transcription PCR (qRT-PCR)
First mandibular molars at PN7 and 11 from Nfic−/− and littermate control mice (n=3 per group) were dissected out, and the apical half was cut with a fine blade (Fine Science Tools, 10050-00) and transferred to RNA stabilization reagent (Qiagen, 76104). RNA was extracted with an RNeasy Mini Kit (Qiagen, 74134) and reverse-transcribed with a QuantiTect Reverse Transcription Kit (Qiagen, 205311). qRT-PCR was performed using the CFX96 Real-Time System (Bio-Rad, iCycler) with EvaGreen Supermix (Bio-Rad, 172-5200). The relative amount of each mRNA transcript was calculated based on a standard curve of cycle thresholds and normalized to Gapdh expression as an internal control. All experiments were repeated three times. Primer sequences were obtained from PrimerBank (http://pga.mgh.harvard.edu/primerbank/). The primer sequences are: Gli1-F: 5′-CCAAGCCAACTTTATGTCAGGG-3′; Gli1-R: 5′-AGCCCGCTTCTTTGTTAATTTGA-3′. Nfic-F: 5′-CCGGCATGAGAAGGACTCTAC-3′; Nfic-R: 5′-TTCTTCACCGGGGATGAGATG-3′. Gapdh-F: 5′-AGGTCGGTGTGAACGGATTTG-3′; Gapdh-R: 5′-TGTAGACCATGTAGTTGAGGTCA-3′.
RNA sequencing
First mandibular molars from PN4 Nfic−/− and sex-matched Nfic+/− littermate control pups were dissected out, and the apical half of the tooth germ was used to extract RNA as described above. cDNA library preparation and sequencing were performed at the Epigenome Center of the University of Southern California. A total of 400 million paired-end reads with 75 cycles were performed on Illumina NextSeq500 equipment for six pairs of samples. Raw reads were trimmed, aligned using TopHat2 with mm10 genome and normalized using RPKM. Differential expression was calculated by selecting transcripts which changed with a significance of P<0.01. RNA-seq data have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE69054.
Chromatin immunoprecipitation (ChIP) assay
The genomic sequence of the 2.5 kb upstream of the transcription start site (TSS) of the mouse Hhip gene was obtained from the University of California, Santa Cruz (UCSC) genome browser as previously described (Iwata et al., 2013). Searches for the putative binding site for NF-I were performed in PROMO (Messeguer et al., 2002) and Matinspector (www.genomatix.de) as previously described (Zhang et al., 2015b). ChIP assays (n=3) were performed using the SimpleChIP kit (Cell Signaling Technology, 9005). For each assay, first molars were dissected out from ten control pups at PN0, minced and cross-linked in 1% formaldehyde for 15 min at room temperature, after which glycine was added to halt the cross-linking. Tissue was then disaggregated using a Dounce grinder and filtered using a 70-µm cell strainer. Nuclei preparation and chromatin digestion were performed according to the manufacturer's protocols. Chromatin fragments were incubated with anti-NF-I antibody (Santa Cruz, sc-5567x; 1:50) (Pjanic et al., 2011) or IgG control antibody. After DNA purification, the following PCR primers were used: NF-I binding site (BS) 1, 5′-gctaggccaaatgtctctgc-3′ and 5′-ggtggaggatgaagctggta-3′; BS 2, 5′-gtctatgcctgccagagagg-3′ and 5′-aggaatctttgcctggcttt-3′. Negative control primers were purchased from Qiagen.
Statistical analysis
Excel 2013 was used for statistical analysis. First, F-tests for variances were performed to confirm equal variances of the samples. Then, we performed two-tailed t-tests of the samples assuming equal variances. P<0.05 was considered statistically significant. No statistical method was used to predetermine sample size.
Acknowledgements
We are grateful to Dr Julie Mayo for critical reading of the manuscript. We thank Dr Andrew McMahon for supplying the Hhip cDNA plasmid. We thank Dr Vasu Punj for GEO submission. We thank the Norris Medical Library Bioinformatics Service at the University of Southern California for assisting with sequencing data analysis. The bioinformatics software and computing resources used in the analysis are funded by the USC Office of Research and the Norris Medical Library.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Y.L. and Y.C. designed the study. Y.L. carried out most of the experiments and analyzed the data. J.F. participated in the experiment. J.L. and H.Z. participated in mice breeding. T.-V.H. contributed to micro-CT scanning. Y.L. and Y.C. co-wrote the paper.
Funding
This study was supported by grants from the National Institute of Dental and Craniofacial Research of the National Institutes of Health (NIH) [R01 DE022503, R37 DE012711 and U01 DE024421 to Y.C.]. Deposited in PMC for release after 12 months.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.127068/-/DC1
References
- Bae C.-H., Kim T.-H., Chu J.-Y. and Cho E.-S. (2013a). New population of odontoblasts responsible for tooth root formation. Gene Expr. Patterns 13, 197-202. 10.1016/j.gep.2013.04.001 [DOI] [PubMed] [Google Scholar]
- Bae C. H., Lee J. Y., Kim T. H., Baek J. A., Lee J. C., Yang X., Taketo M. M., Jiang R. and Cho E. S. (2013b). Excessive Wnt/beta-catenin signaling disturbs tooth-root formation. J. Periodontal. Res. 48, 405-410. 10.1111/jre.12018 [DOI] [PubMed] [Google Scholar]
- Bai C. B., Stephen D. and Joyner A. L. (2004). All mouse ventral spinal cord patterning by hedgehog is Gli dependent and involves an activator function of Gli3. Dev. Cell 6, 103-115. 10.1016/S1534-5807(03)00394-0 [DOI] [PubMed] [Google Scholar]
- Briscoe J. and Thérond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 418-431. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Chai Y., Jiang X., Ito Y., Bringas P. Jr., Han J., Rowitch D. H., Soriano P., McMahon A. P. and Sucov H. M. (2000). Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development 127, 1671-1679. [DOI] [PubMed] [Google Scholar]
- Cho E.-S., Lim S.-S., Hwang J.-W. and Lee J.-C. (2012). Constitutive activation of smoothened leads to impaired developments of postnatal bone in mice. Mol. Cells 34, 399-405. 10.1007/s10059-012-0186-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P.-T. and McMahon A. P. (1999). Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617-621. 10.1038/17611 [DOI] [PubMed] [Google Scholar]
- Chuang P.-T., Kawcak T. and McMahon A. P. (2003). Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 17, 342-347. 10.1101/gad.1026303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule H. R., Lewis P., Bei M., Maas R. and McMahon A. P. (2000). Sonic hedgehog regulates growth and morphogenesis of the tooth. Development 127, 4775-4785. [DOI] [PubMed] [Google Scholar]
- Dessaud E., McMahon A. P. and Briscoe J. (2008). Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development 135, 2489-2503. 10.1242/dev.009324 [DOI] [PubMed] [Google Scholar]
- Gao Y., Yang G., Weng T., Du J., Wang X., Zhou J., Wang S. and Yang X. (2009). Disruption of Smad4 in odontoblasts causes multiple keratocystic odontogenic tumors and tooth malformation in mice. Mol. Cell. Biol. 29, 5941-5951. 10.1128/MCB.00706-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritli-Linde A., Bei M., Maas R., Zhang X. M., Linde A. and McMahon A. P. (2002). Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development 129, 5323-5337. 10.1242/dev.00100 [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M. (2000). Roles of the NFI/CTF gene family in transcription and development. Gene 249, 31-45. 10.1016/S0378-1119(00)00140-2 [DOI] [PubMed] [Google Scholar]
- Gronostajski R. M., Adhya S., Nagata K., Guggenheimer R. A. and Hurwitz J. (1985). Site-specific DNA binding of nuclear factor I: analyses of cellular binding sites. Mol. Cell. Biol. 5, 964-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle Z., Mo R., Hui C. C. and Sharpe P. T. (1998). The Shh signalling pathway in tooth development: defects in Gli2 and Gli3 mutants. Development 125, 2803-2811. [DOI] [PubMed] [Google Scholar]
- Harfe B. D., Scherz P. J., Nissim S., Tian H., McMahon A. P. and Tabin C. J. (2004). Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell 118, 517-528. 10.1016/j.cell.2004.07.024 [DOI] [PubMed] [Google Scholar]
- Huang X., Bringas P. Jr., Slavkin H. C. and Chai Y. (2009). Fate of HERS during tooth root development. Dev. Biol. 334, 22-30. 10.1016/j.ydbio.2009.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Xu X., Bringas P. Jr., Hung Y. P. and Chai Y. (2010). Smad4-Shh-Nfic signaling cascade-mediated epithelial-mesenchymal interaction is crucial in regulating tooth root development. J. Bone Miner. Res. 25, 1167-1178. 10.1359/jbmr.091103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata J.-i., Suzuki A., Pelikan R. C., Ho T.-V., Sanchez-Lara P. A., Urata M., Dixon M. J. and Chai Y. (2013). Smad4-Irf6 genetic interaction and TGFbeta-mediated IRF6 signaling cascade are crucial for palatal fusion in mice. Development 140, 1220-1230. 10.1242/dev.089615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J., Mao J., Tenzen T., Kottmann A. H. and McMahon A. P. (2004). Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 18, 937-951. 10.1101/gad.1190304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M. and Thesleff I. (2012). Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 4, a008425 10.1101/cshperspect.a008425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Seppala M., Zoupa M. and Cobourne M. T. (2007). Hedgehog pathway gene expression during early development of the molar tooth root in the mouse. Gene Expr. Patterns 7, 239-243. 10.1016/j.modgep.2006.10.001 [DOI] [PubMed] [Google Scholar]
- Kim T.-H., Bae C.-H., Jang E.-H., Yoon C.-Y., Bae Y., Ko S.-O., Taketo M. M. and Cho E.-S. (2012). Col1a1-cre mediated activation of beta-catenin leads to aberrant dento-alveolar complex formation. Anat. Cell Biol. 45, 193-202. 10.5115/acb.2012.45.3.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H., Bae C. H., Lee J. C., Ko S. O., Yang X., Jiang R. and Cho E. S. (2013). beta-catenin is required in odontoblasts for tooth root formation. J. Dent. Res. 92, 215-221. 10.1177/0022034512470137 [DOI] [PubMed] [Google Scholar]
- Kolpak A., Zhang J. and Bao Z.-Z. (2005). Sonic hedgehog has a dual effect on the growth of retinal ganglion axons depending on its concentration. J. Neurosci. 25, 3432-3441. 10.1523/JNEUROSCI.4938-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumakami-Sakano M., Otsu K., Fujiwara N. and Harada H. (2014). Regulatory mechanisms of Hertwig's epithelial root sheath formation and anomaly correlated with root length. Exp. Cell Res. 325, 78-82. 10.1016/j.yexcr.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Lee T.-Y., Lee D.-S., Kim H.-M., Ko J. S., Gronostajski R. M., Cho M.-I., Son H.-H. and Park J.-C. (2009). Disruption of Nfic causes dissociation of odontoblasts by interfering with the formation of intercellular junctions and aberrant odontoblast differentiation. J. Histochem. Cytochem. 57, 469-476. 10.1369/jhc.2009.952622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-K., Lee D.-S., Park S.-J., Cho K.-H., Bae H.-S. and Park J.-C. (2014). Nuclear factor I-C (NFIC) regulates dentin sialophosphoprotein (DSPP) and E-cadherin via control of Kruppel-like factor 4 (KLF4) during dentinogenesis. J. Biol. Chem. 289, 28225-28236. 10.1074/jbc.M114.568691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Huang X., Xu X., Mayo J., Bringas P. Jr., Jiang R., Wang S. and Chai Y. (2011). SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development 138, 1977-1989. 10.1242/dev.061341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Feng J., Liu Y., Ho T.-V., Grimes W., Ho H. A., Park S., Wang S. and Chai Y. (2015). BMP-SHH signaling network controls epithelial stem cell fate via regulation of its niche in the developing tooth. Dev. Cell 33, 125-135. 10.1016/j.devcel.2015.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohi M., Tucker A. S. and Sharpe P. T. (2010). Expression of Axin2 indicates a role for canonical Wnt signaling in development of the crown and root during pre- and postnatal tooth development. Dev. Dyn. 239, 160-167. 10.1002/dvdy.22047 [DOI] [PubMed] [Google Scholar]
- Long F., Zhang X. M., Karp S., Yang Y. and McMahon A. P. (2001). Genetic manipulation of hedgehog signaling in the endochondral skeleton reveals a direct role in the regulation of chondrocyte proliferation. Development 128, 5099-5108. [DOI] [PubMed] [Google Scholar]
- Meisterernst M., Gander I., Rogge L. and Winnacker E.-L. (1988). A quantitative analysis of nuclear factor I/DNA interactions. Nucleic Acids Res. 16, 4419-4435. 10.1093/nar/16.10.4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer X., Escudero R., Farre D., Nunez O., Martinez J. and Alba M. M. (2002). PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333-334. 10.1093/bioinformatics/18.2.333 [DOI] [PubMed] [Google Scholar]
- Nakatomi M., Morita I., Eto K. and Ota M. S. (2006). Sonic hedgehog signaling is important in tooth root development. J. Dent. Res. 85, 427-431. 10.1177/154405910608500506 [DOI] [PubMed] [Google Scholar]
- Nakatomi M., Hovorakova M., Gritli-Linde A., Blair H. J., MacArthur K., Peterka M., Lesot H., Peterkova R., Ruiz-Perez V. L., Goodship J. A. et al. (2013). Evc regulates a symmetrical response to Shh signaling in molar development. J. Dent. Res. 92, 222-228. 10.1177/0022034512471826 [DOI] [PubMed] [Google Scholar]
- Ohazama A., Haycraft C. J., Seppala M., Blackburn J., Ghafoor S., Cobourne M., Martinelli D. C., Fan C.-M., Peterkova R., Lesot H. et al. (2009). Primary cilia regulate Shh activity in the control of molar tooth number. Development 136, 897-903. 10.1242/dev.027979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-C., Herr Y., Kim H.-J., Gronostajski R. M. and Cho M.-I. (2007). Nfic gene disruption inhibits differentiation of odontoblasts responsible for root formation and results in formation of short and abnormal roots in mice. J. Periodontol. 78, 1795-1802. 10.1902/jop.2007.060363 [DOI] [PubMed] [Google Scholar]
- Pjanic M., Pjanic P., Schmid C., Ambrosini G., Gaussin A., Plasari G., Mazza C., Bucher P. and Mermod N. (2011). Nuclear factor I revealed as family of promoter binding transcription activators. BMC Genomics 12, 181 10.1186/1471-2164-12-181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakian A., Yang W.-C., Gluhak-Heinrich J., Cui Y., Harris M. A., Villarreal D., Feng J. Q., MacDougall M. and Harris S. E. (2013). Bone morphogenetic protein-2 gene controls tooth root development in coordination with formation of the periodontium. Int. J. Oral Sci. 5, 75-84. 10.1038/ijos.2013.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V. and Briscoe J. (2009). Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb. Perspect. Biol. 1, a002014 10.1101/cshperspect.a002014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoyama W., Liu Y., Yamaza T., Tuan R. S., Wang S., Shi S. and Huang G. T.-J. (2008). Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J. Endod. 34, 166-171. 10.1016/j.joen.2007.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G., Butz K. G., Lyons G. E., Zeichner-David M., Kim H.-J., Cho M.-I. and Gronostajski R. M. (2003). Essential role for NFI-C/CTF transcription-replication factor in tooth root development. Mol. Cell. Biol. 23, 1075-1084. 10.1128/MCB.23.3.1075-1084.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Perkins G., Plachez C., Butz K. G., Yang G., Bachurski C. J., Kinsman S. L., Litwack E. D., Richards L. J. and Gronostajski R. M. (2005). The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol. Cell. Biol. 25, 685-698. 10.1128/MCB.25.2.685-698.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treier M., O'Connell S., Gleiberman A., Price J., Szeto D. P., Burgess R., Chuang P. T., McMahon A. P. and Rosenfeld M. G. (2001). Hedgehog signaling is required for pituitary gland development. Development 128, 377-386. [DOI] [PubMed] [Google Scholar]
- Visbal A. P., LaMarca H. L., Villanueva H., Toneff M. J., Li Y., Rosen J. M. and Lewis M. T. (2011). Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated Smoothened. Dev. Biol. 352, 116-127. 10.1016/j.ydbio.2011.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cox M. K., Coricor G., MacDougall M. and Serra R. (2013). Inactivation of Tgfbr2 in Osterix-Cre expressing dental mesenchyme disrupts molar root formation. Dev. Biol. 382, 27-37. 10.1016/j.ydbio.2013.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Yang G., Wu X., Xie J., Yang X. and Li T. (2013). Disruption of Wnt/beta-catenin signaling in odontoblasts and cementoblasts arrests tooth root development in postnatal mouse teeth. Int. J. Biol. Sci. 9, 228-236. 10.7150/ijbs.5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Jiang Y., Qin C., Liu Y., Ho S. P. and Feng J. Q. (2015a). Essential role of osterix for tooth root but not crown dentin formation. J. Bone Miner. Res. 30, 742-746. 10.1002/jbmr.2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Tian H., Lv P., Wang W., Jia Z., Wang S., Zhou C. and Gao X. (2015b). Transcriptional factor DLX3 promotes the gene expression of enamel matrix proteins during amelogenesis. PLoS ONE 10, e0121288 10.1371/journal.pone.0121288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Feng J., Seidel K., Shi S., Klein O., Sharpe P. and Chai Y. (2014). Secretion of shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell 14, 160-173. 10.1016/j.stem.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Feng J., Ho T.-V., Grimes W., Urata M. and Chai Y. (2015). The suture provides a niche for mesenchymal stem cells of craniofacial bones. Nat. Cell Biol. 17, 386-396. 10.1038/ncb3139 [DOI] [PMC free article] [PubMed] [Google Scholar]