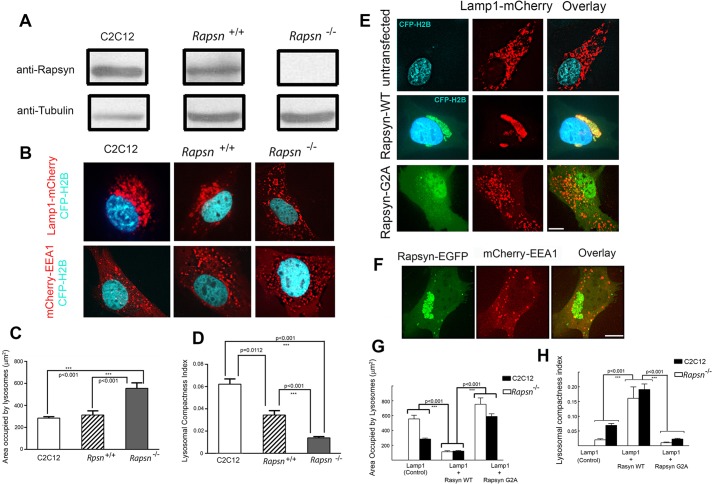
Fig. 7.
The clustering of lysosomes in the juxtanuclear region is impaired in rapsyn-deficient myoblasts, whereas exogenous wild-type rapsyn, but not its non-myristoylated form, rescues the lysosome clustering phenotype. (A) Lysates from C2C12, Rapsn+/+ or Rapsn−/− myoblasts were blotted for endogenous rapsyn using a rabbit monoclonal anti-rapsyn antibody or mouse anti-tubulin for loading control. Endogenous rapsyn was detected in C2C12 myoblasts and Rapsn+/+ myoblasts but not in Rapsn−/− myoblasts. (B) Confocal images of live myoblasts expressing CFP–H2B (to label nuclei) and either Lamp1–mCherry or mCherry–EEA1. The top panel shows representative images of C2C12, Rapsn+/+ or Rapsn−/− myoblasts transfected with lysosomal marker Lamp1–mCherry and CFP–H2B. Although lysosomes are distributed in the juxtanuclear region of C2C12 and Rapsn+/+ myoblasts, they are instead scattered through the entire cytosol in Rapsn−/− myoblasts. The bottom panel shows cells expressing EEA1–mCherry and CFP–H2B. Note that the localization of endosomes is not affected by the loss of endogenous rapsyn as a similar distribution of EEA1 was observed in C2C12, Rapsn+/+ and Rapsn−/− myoblasts, indicating that rapsyn selectively affects the lysosomes clustering. (C) Quantification of the area occupied by lysosomes showing a 2-fold increase in the mean area in Rapsn−/− compared to either Rapsn+/+ or C2C12 myoblasts. P<0.001. Results are mean±s.e.m. (D) Graph showing the quantification of lysosomal compactness index. Note that the mean compactness index in Rapsn−/− is reduced 2.6 times compared to Rapsn+/+ and 4.6 times compared to C2C12 myoblasts. The P values were calculated using Bonferroni test. Results are mean±s.e.m. (E) Representative images of Rapsn−/− myoblasts expressing CFP–H2B (to label nuclei) and Lamp1–mCherry without (top panel), or with wild-type rapsyn–EGFP (middle panel), or Lamp1–mCherry and rapsynG2A–EGFP (lower panel). Note that the scattering of the lysosomes in the absence of rapsyn (top panel) was completely rescued in the presence of exogenous wild-type rapsyn–EGFP as the lysosomes clustered back into the juxtanuclear region. In contrast, expression of rapsynG2A–EGFP was unable to reverse the lysosome scattering defect of Rapsn−/− myoblasts and the lysosomes remained scattered throughout the cytoplasm (lower panel). (F) Representative images of cells showing that the scattered distribution of early endosomes was not affected by overexpression of wild-type rapsyn–EGFP. Scale bars: 10 µm. (G) Quantification of the area occupied by lysosomes in Rapsn−/− myoblasts transfected with Lamp1–mCherry only (untransfected control) or with wild-type rapsyn–EGFP or rapsynG2A–EGFP. Note that in the presence of wild-type rapsyn–EGFP, the area occupied by lysosomes is 5-fold lower than in the absence of rapsyn–EGFP and 6.7-fold lower than in the presence of rapsynG2A–EGFP. For comparison, corresponding data for C2C12 myoblasts are shown side by side. Note that wild-type rapsyn–EGFP rescues the lysosome clustering to the level observed in C2C12 myoblasts. Results are mean±s.e.m. (H) Graph summarizing the quantification of the lysosome compactness index. Note that the index of lysosomal compactness is significantly increased in Rapsn−/− myoblasts transfected with exogenous wild-type rapsyn–EGFP (0.16±0.02, mean±s.e.m., n=10) compared to control Rapsn−/− myoblasts (0.014±0.001, mean±s.e.m., n=29) or Rapsn−/− myoblasts transfected with rapsynG2A–EGFP (0.008±0.001, mean±s.e.m., n=13).