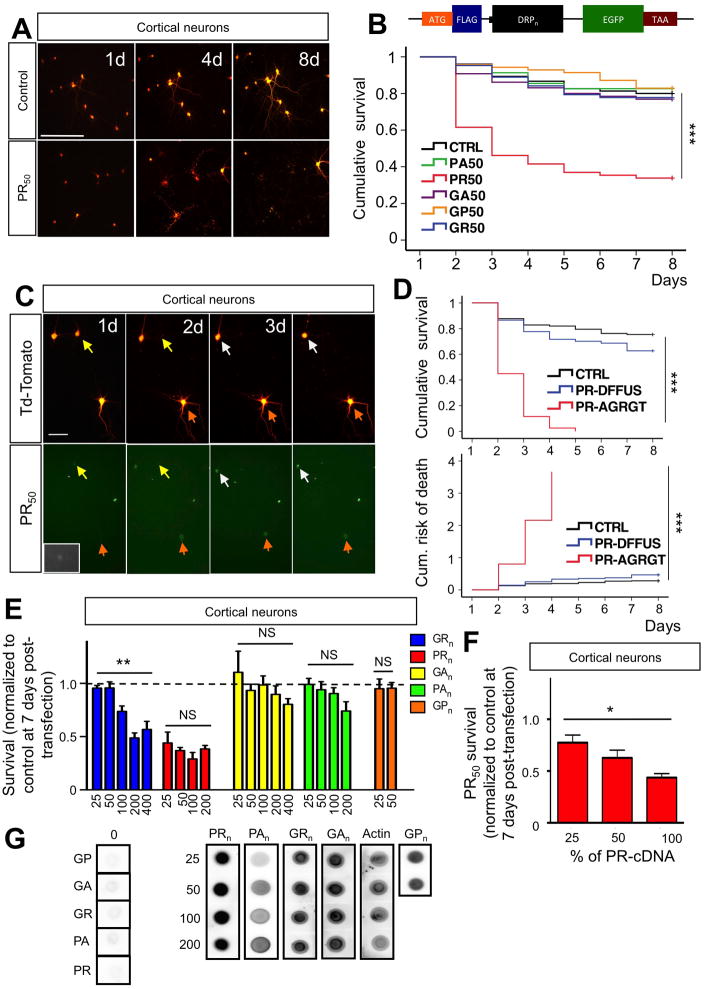
Figure 1. Neurotoxicity of C9RAN poly-dipeptides.
(A) Representative live-cell images of cortical neurons co-transfected at DIV10 with control-GFP + Td-tomato plasmids (4:1) or PR50-GFP + Td-Tomato (4:1). The same neurons were imaged for 9 days (8 days post-transfection) at 24-hour intervals. Calibration bar is 100 μm. (B) Kaplan-Meier survival analysis of cortical neurons co-transfected with constructs encoding different DRPs + Td-Tomato. At least 80 neurons were followed/group; n=5 experiments; ***P<0.001. The insert shows the schematic of the construct. The sequence of the insert is the following: ATG-DYKDDK-KLGR-DPR50-GYRARIHYSSVVEFM-EGFP-TAA. The insert was subcloned into pcDNA3.1 vector and its expression driven by the CMV promoter. The DRP sequence has been constructed with a randomized codon strategy. (C) PR50 aggregates formation preceded neuronal death. Representative images show a group of cortical neurons expressing both PR50 (GFP tagged) and Td-tomato. Neurons in which PR50 formed aggregates underwent cell death within 24–36 hours (yellow arrows, day 1–2; white arrows, day 3–4). Conversely, neurons in which PR50 stayed in a diffused, not aggregated pattern (orange arrow and insets) outlived the neurons with PR aggregates. Calibration bar is 50 μm. (D) Cortical neurons transfected with PR50 that formed aggregates exhibited decreased survival and increased risk of death compared to those that didn’t form detectable PR50 aggregates (***p<0.001). PR50 transfected neurons with no detectable aggregates showed no difference in survival and risk of death compared to control transfected neurons (P = 0.08). (E) Length-dependent toxicity of DRPs in cortical neurons. Poly-GR displayed clear length-dependent toxicity (**P<0.01; one-way ANOVA), whereas the effect of poly-PR already peaked at 25 repeats, displaying no significant length-dependency (one-way ANOVA). (F) Cortical neuron toxicity of PR50 diminished as amount of transfected cDNA was reduced suggesting a dose-dependency in the effect. Maximum amount of cDNA-PR50 transfected was 0.8 μg/well (100%). Total amount of DNA transfected was kept constant across groups (1 μg/well). *P<0.05, **P<0.01, ***P<0.001. (G) NSC34 cells were transfected with indicated constrcuts. Expression levels of different lengths of DRP constructs were assessed using dot blots. Actin was used as a loading control. Comparable expression levels were observed for DRPs of different lengths.