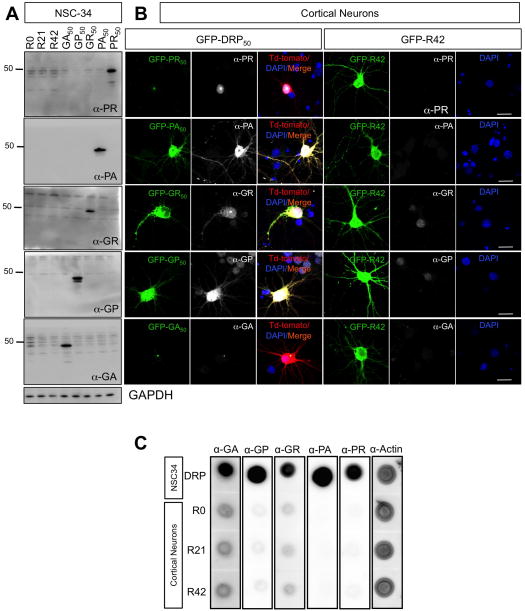
Figure 3. DRPs were not detected in cells transfected with R42 repeat transcript.
(A) NSC34 cells were transfected with the indicated constructs. Efficiency of transfection is ~ 90% for all groups. Homogenates were probed with anti-DRPs antibodies. GAPDH reactivity indicated equal total protein loading across the lanes. Immunoblots showed no detectable DRPs in NSC34 cells transfected with R0, R21 and R42 constructs. (B) Cortical neurons were transfected with the indicated constructs. Immunofluorescence analysis suggested that DRPs did not form in neurons transfected with R42 constructs. Nuclei were stained with DAPI (blue), green represents GFP-DRPs, red represents Td-Tomato, which was cotransfected with GFP-DRP constructs, and white represents the respective DRPs. Calibration bar is 20 μm. (C) Primary cortical neurons were transfected with R0, R21or R42 constructs and collected 72h post-transfection. NSC34 cells transfected with DRP constructs were used as positive controls. Equal amounts of protein were loaded and actin was used as a loading control. Dot blot analysis suggests that no DRPs were generated from cortical neurons expressing R0, R21 and R42.