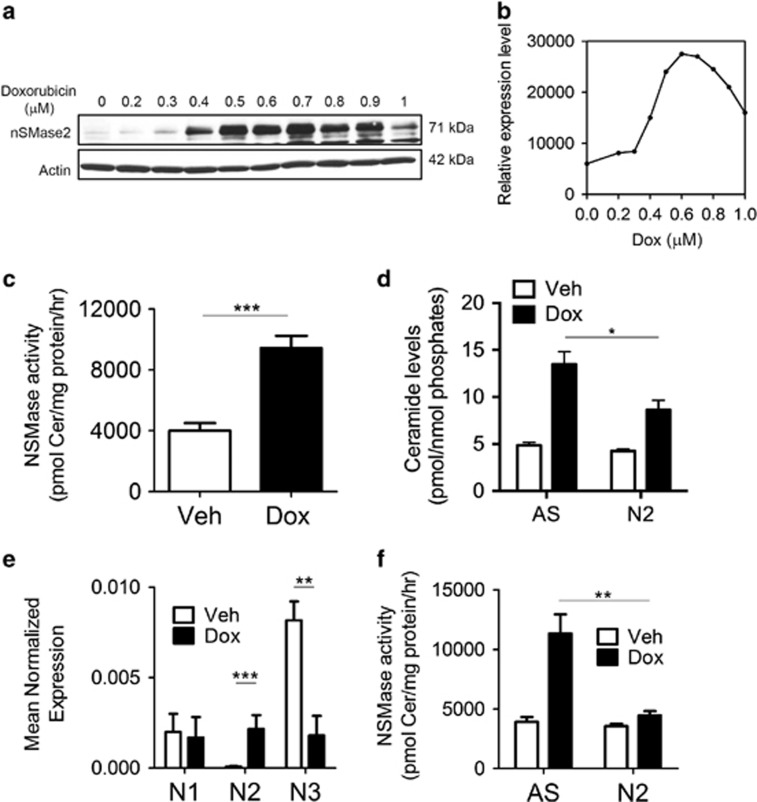
Figure 1.
Doxorubicin induces nSMase2 upregulation and ceramide increase in MCF7 cells. (a) MCF7 cells were plated in 60 mm dishes and treated with either dimethyl sulfoxide (DMSO) or doxorubicin at different doses for 24 h. Cells were collected, lysed and immunoblotted as described under ‘Materials and Methods'. (b) Quantification of nSMase2 induction by ImageJ (NIH, Bethesda, MD, USA) and normalization to actin. (c) MCF7 cells were plated in 60 mm dishes and treated for 24 h with either DMSO or doxorubicin. Cells were collected and in vitro NSMase assay was performed as described under ‘Materials and Methods', ***P<0.001. (d) MCF7 cells were seeded in 60 mm dishes and small interfering RNA (siRNA) was performed to AS (AllStars Negative Control) or nSMase2 (N2) for 24 h, after which either vehicle or doxorubicin were administered and cells were collected 24 h after treatment and analyzed for sphingolipids by liquid chromatography-mass spectrometry (LC/MS) mass spectrometry, *P<0.01. (e) MCF7 cells were lysed 24 h after treatment. RNA was isolated and transformed to cDNA to be quantified by quantitative real-time PCR (qRT-PCR) using primers for nSMase1, nSMase2 and nSMase3 as described, **P<0.01 and ***P<0.001. (f) MCF7 cells were plated in 60 mm dishes. After 24 h, siRNA to AS or nSMase2 (N2) was transfected for 24 h, after which either vehicle or doxorubicin were added. Cells were collected after 24 h, lysed and in vitro sphingomyelinase assay was performed, **P<0.01