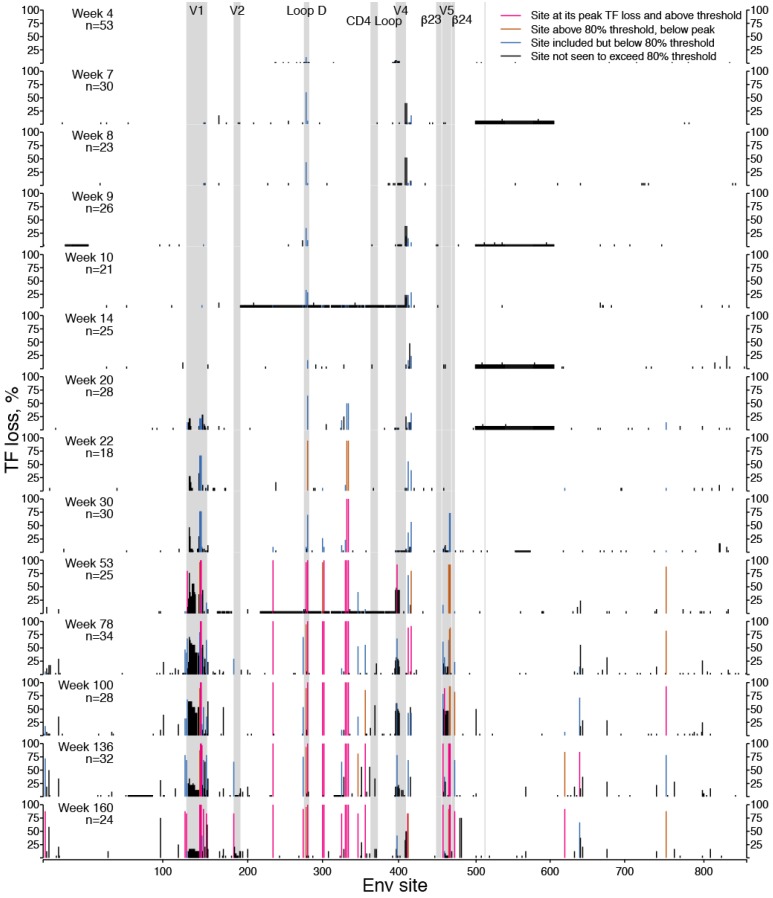
Figure 1.
Loss of ancestral transmitted-founder (TF) amino acids in Envs from CH505. For 953 aligned Env sites spanning the full-length protein, TF loss is proportion of non-TF mutations per time-point sampled from the study participant CH505. TF loss is computed for each of 14 time-points sampled longitudinally, weeks 4 through 160, with the number of Envs sequenced (n) per time-point as shown. Bar colors vary over time to indicate 35 sites with at least 80% TF loss in any time-point, whether at peak TF loss (pink), below peak but above the 80% cutoff (brown), or below 80% (blue). Variable sites that were not selected for further consideration, because they never exceeded 80% TF loss in ant time-point during the study period, are also depicted (black bars). Grey boxes identify variable loops, which contain hypervariable regions that evolve by insertion and deletion, and other gp120 landmarks. A thin grey line marks the boundary between gp120 and gp41. The time of the sample is shown as weeks post infection. Env site numbers indicate HXB2 positions in the CH505 protein alignment, beginning with the signal peptide start codon in column one and ending with the stop codon at position 857.