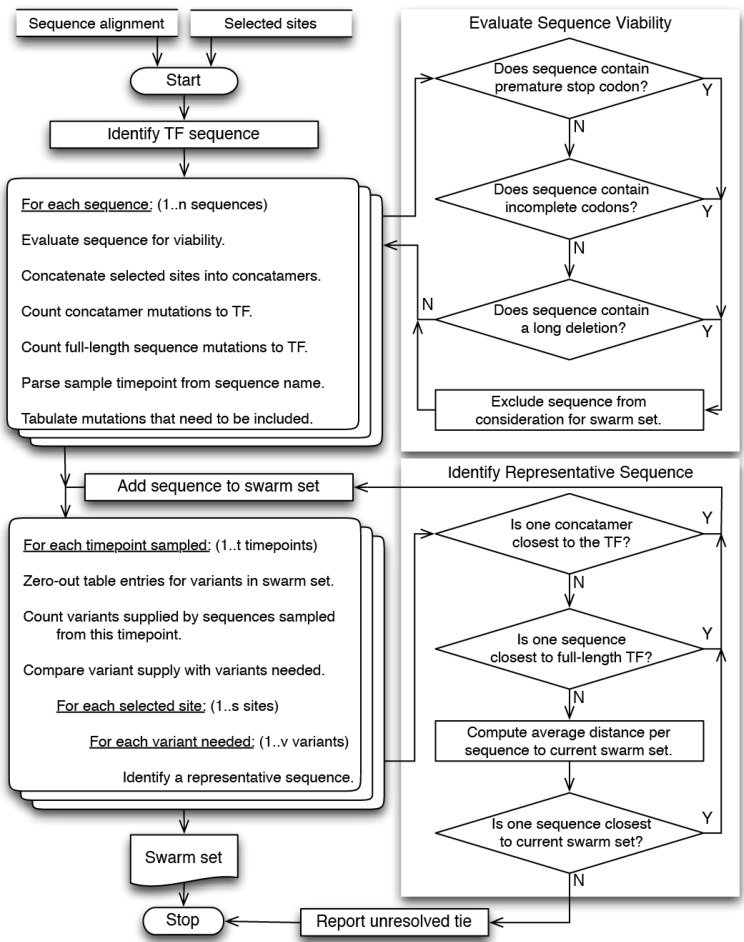
Figure 9.
Swarm-selection algorithm. From a protein sequence alignment and list of selected sites, this approach identifies viable Envs and tabulates mutations in selected sites. The table initially defines which mutations will be represented by the swarm, and subsequently keeps track of which mutations remain to be included. Rare mutations, i.e., mutations detected fewer times than the minimum variant count over the entire sampling period, are disregarded. Selection among multiple sequences that carry a mutation is resolved by minimizing a series of distance criteria, first to minimize Hamming distance (number of mutations, gaps included) to the TF form among selected sites, then distance to the full-length TF sequence, and finally to minimize average distance to sequences in the current swarm set. The selected Env is included in the swarm set, counts in the table of needed mutations are set to zero, to indicate the particular mutation is now covered in the swarm, and iteration continues. This produces a “swarm” of Envs, which represents diversity in selected sites as it developed within the subject, given sampling constraints. Stacked boxes signify iteration. Unresolved ties are reported, though we have not yet encountered them in several large experimental sequence sets we have tested; such an outcome would signal the need for an alternative distance metric or more selection criteria.