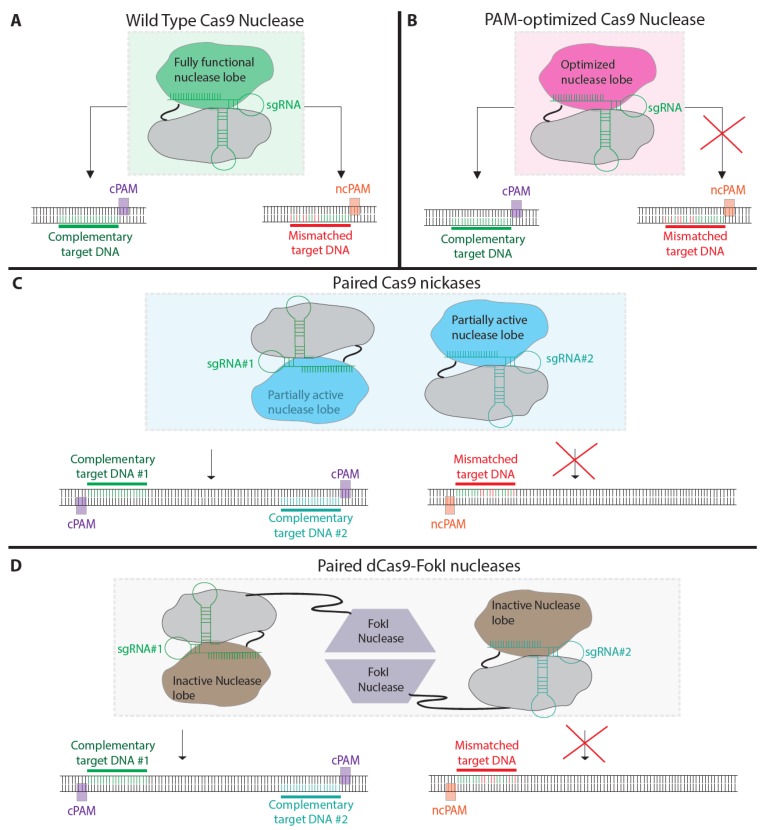
Figure 3.
Modified Cas9 strategies to reduce off-target mutagenesis. (A) Wild type Sp Cas9 nuclease can bind and cleave either sgRNA-complementary (green) or -mismatched (red) DNA with canonical PAM (cPAM) or noncanonical PAM (ncPAM), respectively; (B) PAM-optimized Cas9 nuclease induces cleavage specifically to cPAM complementary DNA; (C) A pair of SpCas9 nickases (i.e., D10A mutant) must bind to two closely spaced DNA sequences on opposite strands in order to induce single strand nicking on each strand and result in full cleavage of the target DNA. The binding of a Cas9 nickase to a non-specific site will induce nicking but not a double strand break; (D) Deactivated Sp dCas9 (i.e., D10A and H840A mutant) fused with a FokI nuclease must bind to two closely spaced DNA sequences in order for dimerization of the FokI nuclease and cleavage to occur. Nonspecific binding at an off-target site will not induce cleavage.