Abstract
Control of HIV replication is a rare immunological event, providing clues to understand the viral control mechanism. CD8+ T-cell responses are crucial for virus control, but it is unclear whether lasting HIV containment can be achieved after establishment of infection. Here, we describe lasting SIV containment in a macaque AIDS model. Analysis of ten rhesus macaques that controlled viremia for 2 years post-infection found accumulation of proviral gag and nef CD8+ T-cell escape mutations in four of them. These four controllers mounted CD8+ T cells targeting Gag, Nef, and other viral proteins at 4 months, suggesting that broadening of CD8+ T-cell targets can be an indicator of the beginning of viral control failure. The remaining six aviremic SIV controllers, however, harbored proviruses without mutations and showed no or little broadening of their CD8+ T-cell responses in the chronic phase. Indeed, three of the latter six exhibiting no change in CD8+ T-cell targets showed gradual decreases in SIV-specific CD8+ T-cell frequencies, implying a concomitant reduction in viral replication. Thus, stability of the breadth of virus-specific CD8+ T-cell responses may represent a status of lasting HIV containment by CD8+ T cells.
Author Summary
CD8+ T-cell responses are crucial for HIV control, but it is unclear whether lasting HIV containment can be achieved after establishment of infection. Several T cell-based vaccine trials have currently shown primary viremia control in macaque AIDS models of simian immunodeficiency virus (SIV) infection, but residual viral replication may occur, followed by accumulation of viral CD8+ T-cell escape mutations, possibly leading to eventual viremia rebound. In the present study, we analyzed ten rhesus macaques that controlled SIV replication without detectable viremia for more than 2 years. Animals were divided into two groups on the basis of proviral genome sequences at 2 years post-infection. Analysis of the first group exhibiting multiple CD8+ T-cell escape mutations indicated that broadening of CD8+ T-cell responses can be an indicator of the beginning of viral control failure. Conversely, analysis of the second group having no mutation suggested that stability of the breadth of virus-specific CD8+ T-cell responses represents a status of lasting HIV containment by CD8+ T cells. Thus, this study presents a model of stable SIV containment, contributing to elucidation of the requisites for lasting HIV control.
Introduction
Human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) infection induces chronic, persistent viral replication leading to AIDS onset in humans and rhesus macaques, respectively. While antiretroviral therapy (ART) has reduced the morbidity and mortality due to HIV, it does not cure infection. Much effort has been made aiming at inducing a functional cure, defined as HIV containment with cessation of ART [1–4]. A current trial of administration with a monoclonal broadly reactive neutralizing antibody under ART showed a longer aviremic period but eventual rebound viremia after ART interruption in rhesus macaques [5,6].
Virus-specific CD8+ T cells exert strong suppressive pressure on HIV/SIV replication [7–11], but fail to control viremia in most infections. Studies of HIV-infected individuals have revealed the association of certain HLA or major histocompatibility complex (MHC) class I genotypes with lower viral loads [12–15]. In the Indian rhesus macaque AIDS model, animals possessing protective MHC alleles such as Mamu-B*08 and Mamu-B*17 tend to show slower disease progression after SIVmac251/SIVmac239 infection [16–18]. CD8+ T-cell responses restricted by these HLA/MHC molecules have been shown to be responsible for HIV/SIV control in most studies [15,19–21]. However, aviremic HIV/SIV control is rare, and even in those with undetectable viremia, residual viral replication can occur and allow accumulation of viral genome mutations resulting in viral escape from CD8+ T-cell recognition, possibly leading to eventual viremia rebound [22–25].
Several prophylactic T cell-based vaccine trials have currently shown primary viremia control in macaque AIDS models [26–29]. However, it is difficult to obtain sterile protection from virus infection by T cell-based vaccines, and whether vaccine-based, primary non-sterile viral control can be stably maintained is debatable. Analysis of those rare cases exhibiting aviremic HIV/SIV control may provide clues to the development of a novel intervention resulting in lasting HIV control.
We previously developed a prophylactic AIDS vaccine using a DNA prime and a boost with a Sendai virus (SeV) vector expressing SIVmac239 Gag (SeV-Gag) [26,30]. Our trial showed vaccine-based control of an SIVmac239 challenge in a group of Burmese rhesus macaques sharing the MHC class I haplotype 90-120-Ia (referred to as A+ animals) [31]. The Mamu-A1*043:01, Mamu-A1*065:01, Mamu-B*061:03, Mamu-B*068:04, and Mamu-B*089:01 alleles have been confirmed in this haplotype [32,33]. Two-thirds of unvaccinated A+ animals showed persistent viremia after SIVmac239 infection, whereas all the A+ animals vaccinated with a DNA prime and an SeV-Gag boost controlled SIV replication without detectable viremia at 2 months post-challenge [31,33]. CD8+ T-cell responses specific for dominant Mamu-A1*043:01 (GenBank accession number AB444869)-restricted Gag206–216 (IINEEAADWDL) and Mamu-A1*065:01 (AB444921)-restricted Gag241–249 (SSVDEQIQW) epitopes are responsible for this vaccine-based SIV control [31]. However, two of these SIV controllers accumulated multiple gag CD8+ T-cell escape mutations and plasma viremia reappeared after 1 year of SIV control [25].
In the present study, we analyzed ten A+ animals that controlled viremia for 2 years after SIVmac239 challenge to determine whether such non-sterile aviremic SIV control can be stably maintained. Four of these ten SIV controllers exhibited an accumulation of proviral CD8+ T-cell escape mutations, indicating the latent viral control failure with preceding broadening of SIV-specific CD8+ T-cell targets. In contrast, the remaining six controllers showed no proviral mutations with little change in CD8+ T-cell targets in the chronic phase. Three of the latter six showed a gradual reduction in SIV-specific CD8+ T-cell frequencies, implying stable non-sterile SIV control with waning viral replication. These results suggest a possible achievement of lasting SIV containment by CD8+ T cells without transition of their targets.
Results
Proviral sequences in SIV controllers
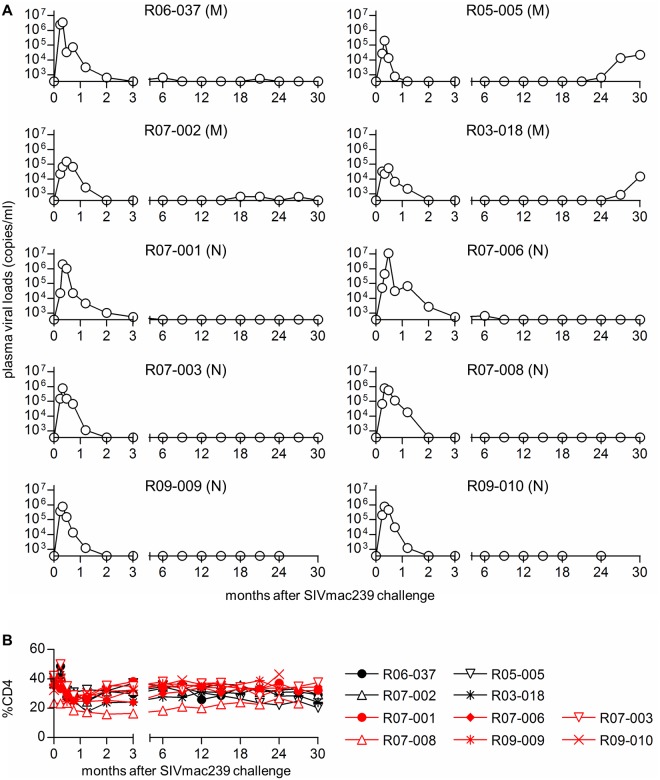
Ten Burmese rhesus macaques possessing the MHC class I haplotype 90-120-Ia that controlled viremia for 2 years after SIVmac239 challenge were studied. In our previous study [34], most A+ animals mounted Gag-specific and Nef-specific CD8+ T-cell responses in primary SIVmac239 infection. Seven 90-120-Ia-associated CD8+ T-cell epitopes, Gag206–216, Gag241–249, Gag367–381, Nef9–19, Nef89–97, Nef193–203, and Vif114–124, in SIVmac239 antigens have been identified, and unvaccinated A+ animals that failed to control viral replication have been shown to select viral escape mutations from all of these epitope-specific CD8+ T cells within a year after SIVmac239 challenge [25,33,34]. Ten SIV controllers used in the present study consisted of two unvaccinated, one sham-vaccinated, and seven vaccinated A+ animals as shown in Table 1. Three macaques (R07-002, R07-003, and R07-008) were immunized with a single Gag241–249-epitope vaccine expressing the Gag241–249 epitope fused with enhanced green fluorescent protein (EGFP) [35], three macaques (R03-018, R09-009, and R09-010) with a single Gag206–216-epitope vaccine expressing the Gag206–216 epitope fused with EGFP [36], and macaque R05-005 with a mixture of both. In these vaccinated animals, plasma viremia became undetectable at 2 months post-infection, whereas unvaccinated controllers showed undetectable viremia after 6 months (Fig 1A). All the animals controlled SIV replication until 2 years, but two of them (R05-005 and R03-018) clearly exhibited reappearance of plasma viremia after that (Fig 1A). Viremia was undetectable at 1 year post-infection in all the ten SIV controllers even by quantitation of viral loads using fivefold-concentrated plasma (Table 2). However, this higher-sensitive assay detected marginal levels of viremia at 2 years post-infection in macaques R05-005 and R03-018 (Table 2). Peripheral CD4+ T lymphocytes were maintained in all of these SIV controllers (Fig 1B).
Table 1. SIV controllers analyzed in this study.
| Macaques | Vaccination a | Virus recovery b |
|---|---|---|
| Group M c | ||
| R06-037 | unvaccinated | positive |
| R05-005 | Gag206–216/241–249 | positive |
| R07-002 | Gag241–249 | positive |
| R03-018 | Gag206–216 | negative |
| Group N c | ||
| R07-001 | unvaccinated | negative |
| R07-006 | control | positive |
| R07-003 | Gag241–249 | negative |
| R07-008 | Gag241–249 | negative |
| R09-009 | Gag206–216 | negative |
| R09-010 | Gag206–216 | negative |
aAnimals were unvaccinated or vaccinated with DNA-prime/SeV-boost expressing EGFP (control), Gag202–216-EGFP and Gag236–251-EGFP (Gag206–216/241–249), Gag236–251-EGFP (Gag241–249), or Gag202–216-EGFP (Gag206–216) before SIVmac239 challenge. All of them controlled SIV replication for more than 2 years.
bpositive, viral gag fragments were detected by RT-PCR amplification from culture supernatant-derived RNAs of PBMCs at 2 years; negative, those were undetectable.
cOn the basis of the data on proviral gag nucleotide sequences at 2 years post-infection, animals were divided into two groups, Group M with multiple mutations and Group N with no mutation.
Fig 1. Plasma viral loads and peripheral %CD4 in SIV controllers.
(A) Plasma viral loads (SIV gag RNA copies/ml plasma) determined as described previously [26]. The lower limit of detection is approximately 4 x 102 copies/ml. On the basis of the data on proviral gag nucleotide sequences at 2 years post-infection, animals were divided into two groups, Group M (M) with multiple CD8+ T-cell escape mutations and Group N (N) with no mutation. (B) Percentage of CD4+ T cells in PBMCs.
Table 2. Plasma viral loads determined by using fivefold-concentrated plasma a .
| Macaques | VL at 1 year pi | VL at 2 years pi |
|---|---|---|
| R06-037 | undetectable | ND |
| R05-005 | undetectable | 3.3 x 102 copies/ml |
| R07-002 | undetectable | undetectable |
| R03-018 | undetectable | 1.3 x 102 copies/ml |
| R07-001 | undetectable | undetectable |
| R07-006 | undetectable | ND |
| R07-003 | undetectable | undetectable |
| R07-008 | undetectable | undetectable |
| R09-009 | undetectable | undetectable |
| R09-010 | undetectable | undetectable |
aAfter centrifugation of 1 ml of plasma at 25,000 x g for 2 hours, 0.8 ml of its supernatant was discarded for fivefold concentration of plasma. The remaining 0.2 ml was subjected to RNA extraction for quantitation of viral loads (VL) [25]. Plasma samples of R06-037 and R07-006 at 2 years post-infection (pi) were unavailable (ND, not determined).
We examined whether viral gag CD8+ T-cell escape mutations accumulated during SIV control. Plasma viral loads were too low for us to recover viral gag cDNA fragments by reverse transcription (RT) and nested PCR amplification from concentrated plasma-derived RNA of these SIV controllers during the chronic phase of infection. We, therefore, analyzed sequences of proviral gag cDNA fragments amplified by nested PCR. Template DNAs were extracted from CD4+ T cells isolated from macaque peripheral blood mononuclear cells (PBMCs) obtained at approximately 2 years post-infection (Fig 2). Four animals (referred to as Group M [Table 1]) had multiple proviral gag mutations, exhibiting the accumulation of CD8+ T-cell escape mutations in the chronic phase. All Group M animals showed selection of mutations in Gag206–216, Gag241–249, and Gag367–381 epitope-coding regions with additional gag mutations. We have previously reported that the L216S (leading to a L-to-S substitution at the 216th amino acid [aa] in Gag) and D205E (D-to-E at the 205th) mutations result in escape from Gag206–216-specific CD8+ T-cell recognition [25,37]. We have also confirmed that D244E (D-to-E at the 244th) results in escape from Gag241–249-specific CD8+ T cells and that A373T (A-to-T at the 373rd) and V375M (V-to-M at the 375th) from Gag367–381-specific CD8+ T cells [25]. In contrast, the remaining six SIV controllers (referred to as Group N [Table 1]) showed no proviral mutations in Gag206–216, Gag241–249, and Gag367–381 epitope-coding regions. No mutation in other gag region was found in Group N animals except for macaque R09-010 having a single M424T mutation. Viral gag cDNA fragments were amplified from culture supernatants of CD4+ T cells derived from three of four Group M animals but not from Group N animals except for macaque R07-006 (Table 1), implying inefficient transcription or replication capacity of the proviruses with no gag mutations in Group N animals.
Fig 2. Dominant non-synonymous mutations in proviral gag in SIV controllers.
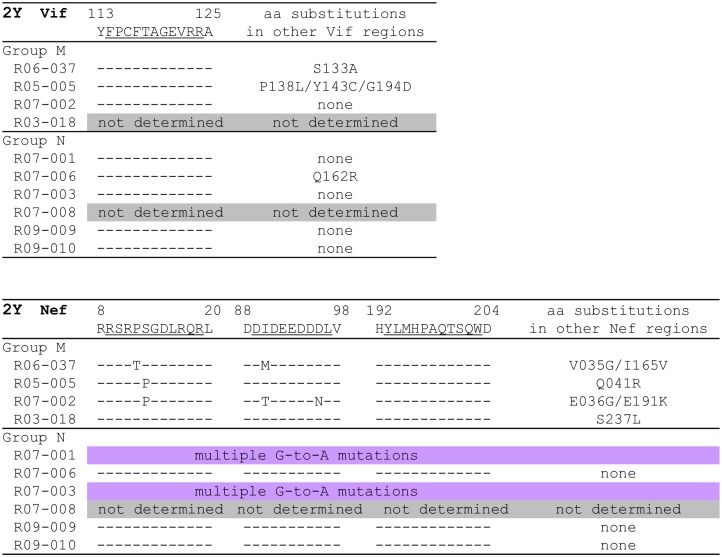
Amino acid substitutions around SIV Gag206–216, Gag241–249, and Gag367–381 epitopes and in other Gag regions approximately 2 months (2M, top), 1 year (1Y, middle), and 2 years (2Y, bottom) after SIVmac239 challenge are shown. Most of the proviral gag fragments were amplified from CD4+ T cells isolated from PBMCs, while those at 2 years in macaques R06-037, R05-005, R07-001, and R07-006 were from cultured CD4+ T cells due to limitation of available cell numbers. Mutant sequences shown were completely dominant (i.e., wild-type sequences were undetectable at the residues showing mutant sequences) except for the L216S mutation (the ratio of wild type/mutant: 2/5) in macaque R03-018 at 1 year post-infection. No subdominant mutation was detected.
We also analyzed proviral gag sequences in PBMCs obtained at 2 months and 1 year after SIVmac239 challenge (Fig 2). In contrast to the results obtained at 2 years post-infection, no proviral gag mutations were observed in Group M or Group N animals, except macaque R03-018 (Group M) having a single L216S mutation at 1 year. Thus, Group M macaques accumulated proviruses with multiple CD8+ T-cell escape mutations later than 1 year post-infection and had replication-competent viruses that could be recovered from PBMC culture at 2 years.
We then examined proviral vif and nef sequences at 2 years in these SIV controllers (Fig 3). Proviral vif cDNA fragments were amplified from three of four Group M and five of six Group N animals. Proviral vif mutations were found in two of the three Group M but only one of the five Group N. Neither Group M nor N had mutations in the Vif114–124 CD8+ T-cell epitope-coding region.
Fig 3. Dominant non-synonymous mutations in proviral vif and nef in SIV controllers.
In the upper panel, amino acid substitutions around SIV Vif114–124 epitope and in other Vif regions approximately 2 years after SIVmac239 challenge are shown. In the lower panel, amino acid substitutions around SIV Nef9–19, Nef89–97, and Nef193–203 epitopes and in other Nef regions approximately 2 years after SIVmac239 challenge are shown. Sequences of vif in macaques R03-018 and R07-008 and nef in macaques R07-008 were not determined because these cDNA fragments could not be amplified. Macaques R07-001 and R07-003 had multiple G-to-A mutations in nef (See S1 Fig). Mutant sequences shown were completely dominant except for vif P138L (the ratio of wild type/mutant: 1/2) in R05-005, vif Q162R (1/1) in R07-006, nef P012T (1/10) in R06-037, and nef S013P (3/10), I090T (1/5), D096N (1/5), and E191K (1/5) in R07-002. In addition, subdominant nef mutations resulting in P012S (5/2) and G044E (5/2) were detected in macaque R07-002, while the wild-type sequences were dominant at these positions.
Group M animals had multiple proviral nef mutations, exhibiting accumulation of CD8+ T-cell escape mutations during the chronic phase of infection. Mutations in the Nef9–19- and Nef89–97-coding regions were selected in three and two of four Group M animals, respectively. We have previously reported that the P12T (P-to-T at the 12th aa in Nef) and S13P (S-to-P at the 13th) were Nef9–19-specific CD8+ T-cell escape mutations [34]. In the Nef193–203-coding region, however, no mutation was observed even in Group M animals. In contrast, Group N controllers showed no mutation in proviral Nef9–19- Nef89–97- and Nef193–203-coding regions, while macaques R07-001 and R07-003 had multiple G-to-A mutations (S1 Fig), possibly due to the effect of the APOBEC3 family [38–41]. Thus, we found two groups of A+ SIV controllers, Group M accumulating CD8+ T-cell escape mutations and Group N having no dominant CD8+ T-cell escape mutations in proviruses.
SIV antigen-specific CD8+ T-cell responses in SIV controllers
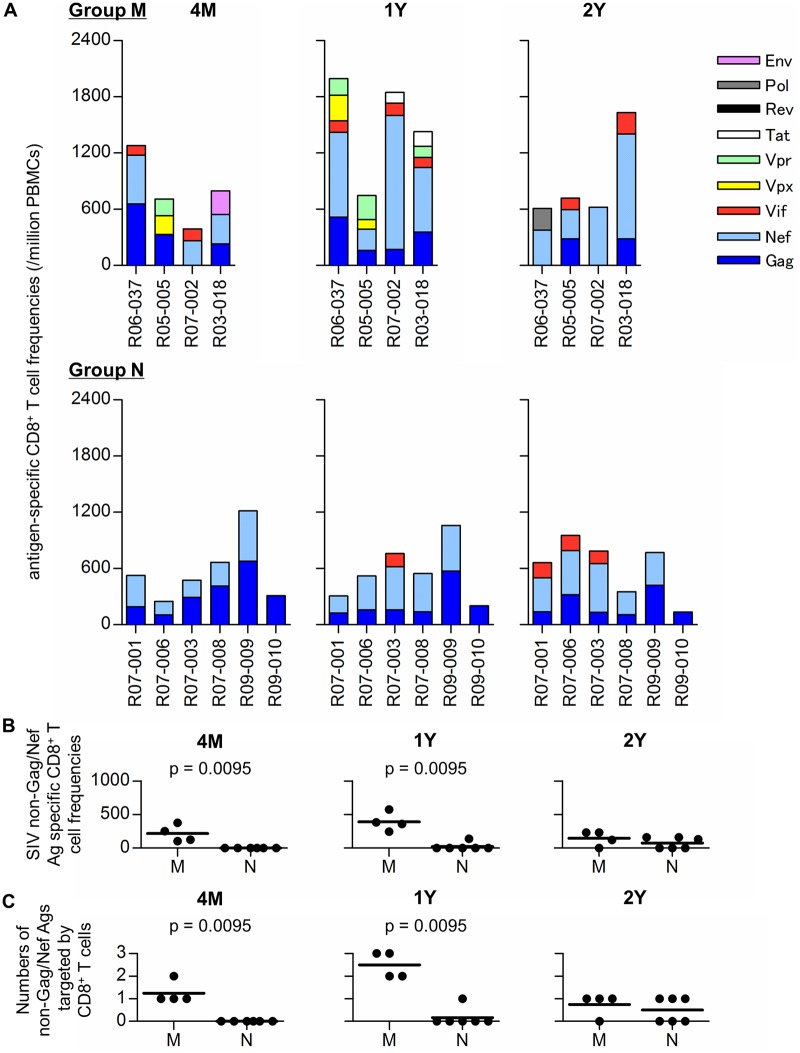
Next, we examined individual SIV antigen-specific CD8+ T-cell responses in our SIV controllers by using panels of overlapping peptides spanning the entire SIVmac239 Gag, Pol, Vif, Vpx, Vpr, Tat, Rev, Env, and Nef amino acid sequences (Fig 4A). No significant difference in SIV whole antigen-specific CD8+ T-cell frequencies (sum of individual antigen specific CD8+ T-cell frequencies) was observed between Groups M and N at 4 months post-infection. However, the frequencies in Group M were significantly higher than in Group N at 1 year post-infection (p = 0.0381 by Mann-Whitney U-test). All the Group M animals had higher frequencies at 1 year than at 4 months, but Group N showed no or minimal increase in the whole antigen-specific CD8+ T-cell frequencies. In particular, three Group N controllers, R07-008, R09-009, and R09-010, exhibited gradual decreases in the whole antigen-specific CD8+ T-cell frequencies from 4 months to 2 years following challenge (Fig 4A).
Fig 4. SIV antigen-specific CD8+ T-cell responses in SIV controllers.
(A) Frequencies of CD8+ T cells specific for Gag, Nef, Vif, Vpx, Vpr, Tat, Rev, Pol, and Env in Group M (upper panels) and Group N (lower) at 4 months (4M), 1 year (1Y), and 2 years (2Y) post-infection. Responses were measured by detection of antigen-specific IFN-γ induction using panels of overlapping peptides spanning the entire SIVmac239 Gag, Nef, Vif, Vpx, Vpr, Tat, Rev, Pol, and Env amino acid sequences, respectively. (B) Comparisons of CD8+ T-cell frequencies specific for SIV antigens other than Gag and Nef at 4M, 1Y, and 2Y between Groups M and N. Group M had significantly higher frequencies of SIV non-Gag/Nef antigen-specific CD8+ T cells at 4M (p = 0.0095 by Mann-Whitney U-test) and 1Y (p = 0.0095). (C) Comparisons of the numbers of CD8+ T cell-targeted SIV antigens other than Gag and Nef at 4M, 1Y, and 2Y between Groups M and N. The numbers were significantly higher in Group M at 4M (p = 0.0095 by Mann-Whitney U-test) and 1Y (p = 0.0095).
All the A+ SIV controllers induced predominant Gag- and/or Nef-specific CD8+ T-cell responses at 4 months after SIVmac239 infection (Fig 4A). Group M animals elicited additional CD8+ T-cell responses directed against SIV antigens other than Gag and Nef. These SIV non-Gag/Nef antigen-specific CD8+ T-cell frequencies were significantly higher in Group M than those in Group N at 4 months (p = 0.0095 by Mann-Whitney U-test) and 1 year (p = 0.0095) (Fig 4B). Indeed, the numbers of SIV non-Gag/Nef antigens targeted by CD8+ T cells were significantly higher in Group M than those in Group N at 4 months (p = 0.0095 by Mann-Whitney U-test) and 1 year (p = 0.0095) (Fig 4C). All the Group M animals showed increase in the numbers of SIV antigens targeted by CD8+ T cells at 1 year compared to those at 4 months. In particular, SIV non-Gag antigen-specific CD8+ T-cell frequencies were significantly higher in Group M at 1 year (p = 0.0087). These results indicate the broadening of the CD8+ T-cell response in Group M.
All four animals in Group M mounted CD8+ T-cell responses specific for SIV non-Gag/Nef antigens, which were undetectable in all Group N animals at 4 months post-infection. Also at 1 year, all of the animals in Group M showed CD8+ T-cell responses specific for several targets besides the Gag and Nef antigens. In contrast, only Gag-specific and Nef-specific CD8+ T-cell responses were observed in Group N animals except for macaque R07-003 which had detectable Vif-specific CD8+ T-cell responses. At 2 years post-infection, Vif-specific CD8+ T-cell responses were detected in three macaques in Group N, while the remaining three showed Gag-specific and Nef-specific CD8+ T-cell responses only. Importantly, the latter three macaques (R07-008, R09-009, and R09-010) exhibited gradual decreases in these Gag/Nef-specific CD8+ T-cell responses in the chronic phase, possibly reflecting the absence of measurable viral replication.
SIV epitope-specific CD8+ T-cell responses in SIV controllers
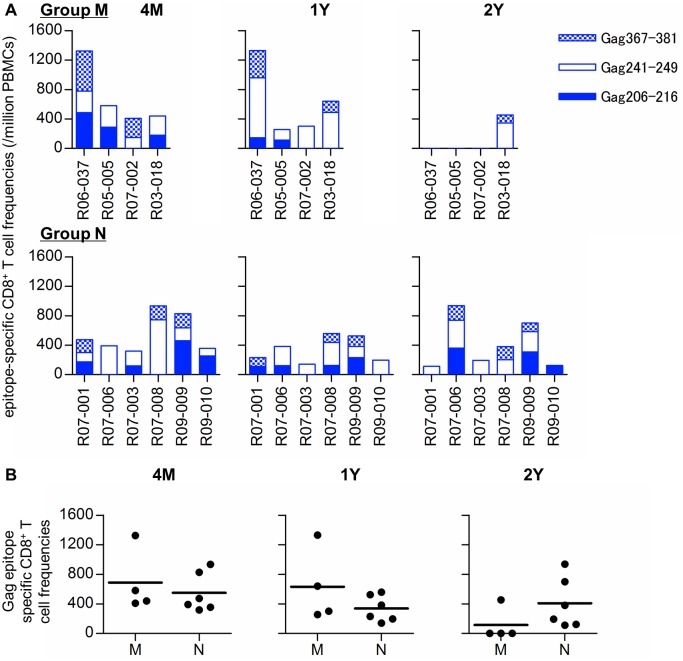
We further examined CD8+ T-cell responses specific for 90-120-Ia-associated Gag epitopes; Gag206–216, Gag241–249, and Gag367–381, in the ten SIV controllers (Fig 5A). The sum of these Gag epitope-specific CD8+ T-cell frequencies was similar between Groups M and N (Fig 5B). Induction of CD8+ T-cell responses directed against both dominant Gag206–216 and Gag241–249 epitopes were confirmed in all the animals in previous analyses at weeks 2 and 12 post-infection [35,36]. At 4 months, all had detectable Gag241–249-specific CD8+ T cells, although Gag206–216-specific CD8+ T-cell responses were undetectable in three animals. However, at 2 years post-infection, the Gag241–249-specific CD8+ T-cell responses became undetectable in three of four Group M animals but were maintained in Group N animals except for macaque R09-010 which exhibited Gag206–216-specific CD8+ T-cell responses.
Fig 5. Gag206–216, Gag241–249, and Gag367–381 epitope-specific CD8+ T-cell responses in SIV controllers.
(A) Frequencies of CD8+ T cells specific for SIV Gag206–216, Gag241–249, and Gag367–381 epitopes in Group M (upper panels) and Group N (lower) at 4 months (4M), 1 year (1Y), and 2 years (2Y) post-infection. (B) Comparisons of the sum of Gag206–216-, Gag241–249-, and Gag367–381-specific CD8+ T-cell frequencies at 4M, 1Y, and 2Y between Groups M and N. No significant difference was observed between the groups.
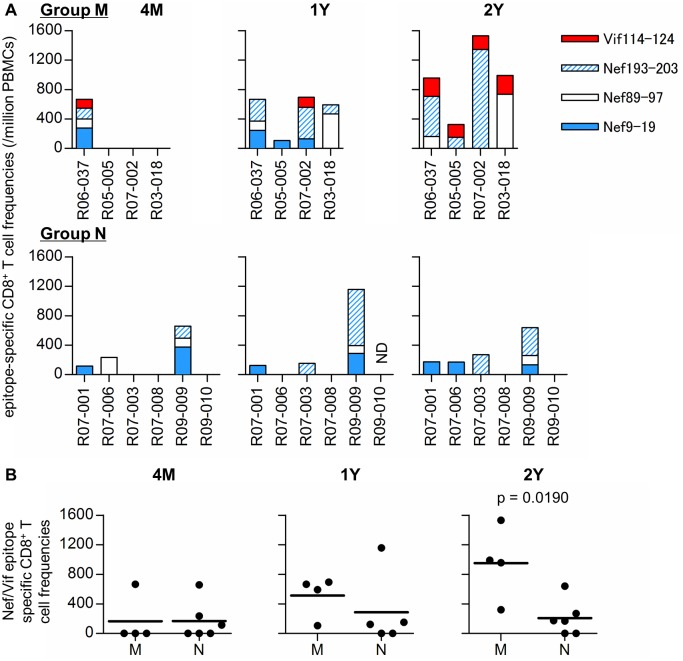
We also examined CD8+ T-cell responses specific for 90-120-Ia-associated Nef9–19, Nef89–97, Nef193–203, and Vif114–124 epitopes in the SIV controllers (Fig 6A). In most of the animals immunized with Gag241–249-epitope and/or Gag206–216-epitope vaccines (except for macaque R09-009), these Gag epitope-specific CD8+ T-cell responses were predominant but Nef/Vif epitope-specific CD8+ T-cell responses were undetectable at 4 months post-challenge. At 1 and 2 years post-infection, differences in these Nef/Vif epitope-specific CD8+ T-cell responses between Groups M and N became evident. All the Group M animals mounted Nef9–19- Nef89–97-, or Nef193–203-specific CD8+ T-cell responses at 1 year and Vif114–124-specific CD8+ T-cell responses at 2 years post-infection. In contrast, none of the Group N animals elicited Vif114–124-specific CD8+ T-cell responses. In two of them, macaques R07-008 and R09-010, even Nef9–19- Nef89–97-, and Nef193–203-specific CD8+ T cells were undetectable. Indeed, the sum of Nef9–19- Nef89–97-, Nef193–203-, and Vif114–124-specific CD8+ T-cell frequencies in Group M was significantly higher than in Group N at 2 years (p = 0.0190 by Mann-Whitney U-test) (Fig 6B). Thus, Group M animals mounted Nef and Vif epitope-specific CD8+ T-cell responses in the chronic phase of infection, whereas such broadening of CD8+ T-cell responses was unclear and Gag epitope-specific CD8+ T cells were maintained in Group N. In particular, in three of six Group N macaques, R07-008, R09-009, and R09-010, comparison between 4 months and 2 years post-challenge showed no change in CD8+ T-cell target epitopes.
Fig 6. Nef9–19, Nef89–97, Nef193–203, and Vif114–124 epitope-specific CD8+ T-cell responses in SIV controllers.
(A) Frequencies of CD8+ T cells specific for SIV Nef9–19, Nef89–97, Nef193–203, and Vif114–124 epitopes in Group M (upper panels) and Group N (lower) at 4 months (4M), 1 year (1Y), and 2 years (2Y) post-infection. (B) Comparisons of the sum of Nef9–19-, Nef89–97-, Nef193–203-, and Vif114–124-specific CD8+ T-cell frequencies at 4M, 1Y, and 2Y between Groups M and N. The sum of CD8+ T-cell frequencies specific for these epitopes in Group M was significantly higher compared to Group N at 2Y post-infection (p = 0.0190 by Mann-Whitney U-test).
CD8+ cell-depletion in a Group N controller
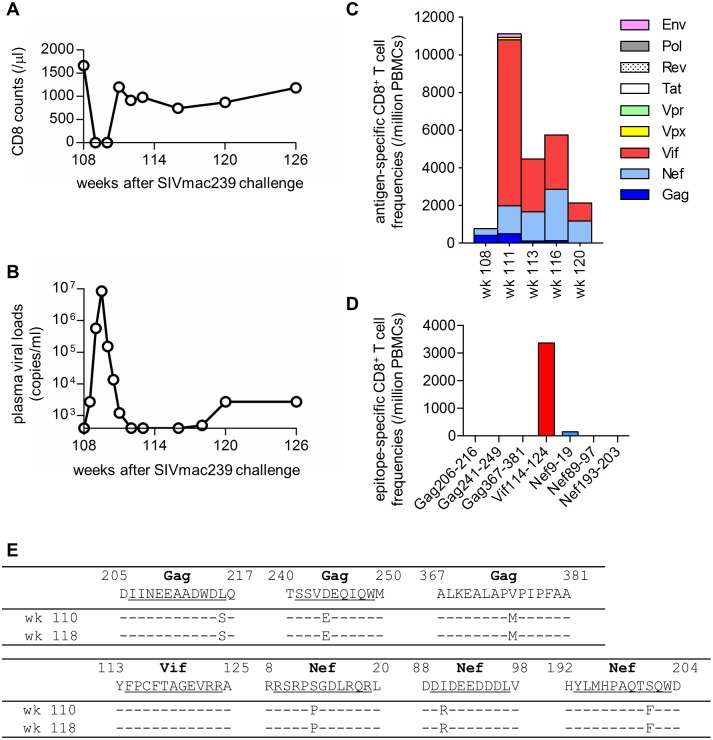
Three of six Group N controllers, R07-008, R09-009, and R09-010, showed gradual decreases and stable breadth in the SIV antigen-specific CD8+ T-cell responses from 4 months to 2 years post-infection. To confirm involvement of the CD8+ T-cell responses in this stable SIV control, we administered an anti-CD8 antibody to deplete the CD8+ cells in macaque R09-009 2 years following challenge. Peripheral CD8+ T cells were undetectable for approximately 2 weeks from day 3 after the initial anti-CD8 antibody administration at week 108 post-infection and became detectable at week 111 (Fig 7A). Plasma viremia was detectable for approximately 3 weeks from day 3 after the initial anti-CD8 antibody administration and became undetectable at week 112 (Fig 7B). This transient reappearance of plasma viremia concomitant with CD8+ cell depletion supports the notion that CD8+ T-cell responses are crucial for the stable SIV containment.
Fig 7. Virological and immunological analyses in macaque R09-009 following CD8+ cell depletion.
(A) Changes in peripheral CD8+ T-cell counts after the initial anti-CD8 antibody administration. Group N macaque, R09-009, was administered anti-CD8 antibody at week 108 post-infection and on days 3, 7, and 10 after the first administration. (B) Changes in plasma viral loads. (C) Changes in CD8+ T-cell responses specific for SIV Gag, Nef, Vif, Vpx, Vpr, Tat, Rev, Pol, and Env. (D) CD8+ T-cell responses specific for SIV Gag206–216, Gag241–249, Gag367–381, Vif114–124, Nef9–19, Nef89–97, and Nef193–203 epitopes at week 113 post-infection. (E) Dominant non-synonymous mutations in plasma viral cDNA regions encoding Gag, Vif, and Nef epitopes. Viral gag, vif, and nef cDNA fragments were amplified from plasma RNA obtained at weeks 110 and 118 post-infection. Amino acid substitutions around SIV Gag206–216, Gag241–249, Gag367–381, Vif114–124, Nef9–19, Nef89–97, and Nef193–203 epitopes are shown.
Macaque R09-009 showed Gag- and Nef-specific but not non-Gag/Nef antigen-specific CD8+ T-cell responses before the anti-CD8 antibody administration at week 108 post-infection (Fig 4A). Interestingly, however, this animal mounted high magnitude Vif-specific CD8+ T-cell responses in the CD8+ T-cell recovery phase from week 111 post-infection (Fig 7C). Analysis at week 115 detected no or poor Gag/Nef epitope-specific CD8+ T cells but dominant Vif114–124 epitope-specific CD8+ T-cell responses (Fig 7D). The plasma-derived viral genome cDNAs at week 110 post-infection had mutations L216S in the Gag206–216 epitope-coding region, D244E in Gag241–249, V375M in Gag367–381, S13P in Nef9–19, I90R in Nef89–97, and S201F in Nef193–203 (Fig 7E). However, no mutation was detected in the Vif114–124 epitope-coding region. Plasma viremia re-appeared at week 118, and the viruses at week 118 showed the similar pattern of sequences (Fig 7E). Thus, even in lasting aviremic SIV controllers, replication-competent viruses with multiple CD8+ T-cell escape mutations may exist.
Discussion
In the present study, we examined ten rhesus macaques that controlled SIV replication without detectable viremia for more than 2 years after infection. Four of these aviremic SIV controllers, Group M, exhibited proviruses with multiple proviral CD8+ T-cell escape mutations in the chronic phase, whereas the remaining six, Group N, had no proviral CD8+ T-cell escape mutations. Three of six Group N animals showed a decline in SIV-specific CD8+ T-cell frequencies, implying lasting non-sterile SIV control with a concomitant reduction in viral replication. Although the size and character of virus reservoirs may be different from those under ART [42,43], a rhesus cytomegalovirus (rhCMV) vector vaccine trial indicated lasting, non-sterile SIV control by persistent rhCMV-induced CD8+ T-cell responses, possibly resulting in virus clearance [44]. Our results suggest possible achievement of lasting SIV control by CD8+ T cells without persistent exogenous stimulation. We were unable to find the genetic determinants for the difference in the Groups M and N by analyses of their second MHC class I haplotypes (Table 3), but this study presents a model of SIV containment, contributing to elucidation of the requisites for lasting, non-sterile HIV control.
Table 3. Alleles in the second MHC-I haplotypes in macaques a .
| Macaques | Alleles |
|---|---|
| R06-037 | A1*052:01, A2*05:13, B*089:02/03 |
| R05-005 | A1*105:02, B*056:03, B*066:01 |
| R07-002 | A1*066:01, B*021:02 |
| R03-018 | A1*018:08, A2*05:31, B*001:01, B*007:02/03 |
| R07-001 | A1*032:02, B*066:01 |
| R07-006 | A1*032:02 |
| R07-003 | A1*066:01, B*003:01, B*005:01, B*007:02/03, B*015:04 |
| R07-008 | A1*004:01, A1*018:01, A2*05:31, B*043:01 |
| R09-009 | A1*107:01, B*046:03 |
| R09-010 | B*063:02, B*066:01 |
aDetected alleles not included in the first MHC-I haplotype 90-120-Ia are shown.
This study analyzed SIV controllers possessing the MHC-I haplotype 90-120-Ia. In our previous study [31], all the A+ animals vaccinated with a DNA prime and an SeV-Gag boost controlled replication of the wild-type SIVmac239, whereas those that received the same vaccine regimen failed to control a challenge with SIV carrying Gag206–216- and Gag241–249-specific CD8+ T-cell escape mutations. Thus, these A+ animals are useful to examine the mechanism of SIV control by CD8+ T cells, and the present study using these SIV controllers may provide a clue to understand the mechanism for lasting SIV control. Analysis of CD8+ T cell-mediated SIV control needs animals sharing MHC-I genotypes like A+ macaques. If we could accumulate long-lasting aviremic SIV controllers sharing other MHC-I alleles, although not easy, it would contribute to our further understanding of viral control mechanism.
Gag206–216- and Gag241–249-specific CD8+ T-cell responses played a central role in primary viral control in these A+ SIV controllers as shown previously [31, 35,36]. In most vaccine-based controllers, a viral CD8+ T-cell escape GagL216S mutation was rapidly selected and plasma viremia became undetectable after 5 weeks of infection. However, the wild-type sequence but not this CD8+ T-cell escape mutation was predominant in PBMC-derived proviruses at 2 months post-infection, reflecting containment of the rapidly-selected mutant viruses. The data infer that the detected proviruses with wild-type gag were maintained without eradication because of inefficient replication. Indeed, no virus was recovered from PBMC cultures in most Group N animals (Table 1). How these proviruses became non-functional remains unclear [45,46], but two Group N animals showed multiple G-to-A mutations in proviral nef (S1 Fig), possibly reflecting an effect of the APOBEC3 family [38–41].
Two years after infection, animals in Group M showed multiple CD8+ T-cell escape mutations in proviral gag and nef, suggesting replication of viruses carrying multiple mutations. In our previous study [33], all the four MHC-I haplotype 90-120-Ia-positive macaques that failed to control SIVmac239 replication showed CD8+ T-cell responses targeting SIV non-Gag/Nef as well as Gag/Nef antigens at 3 months and/or 1 year post-infection. In all of these animals, analysis of plasma RNA-derived viral genome sequences at 1 year post-infection found predominant nonsynonymous mutations in all the seven regions encoding SIV Gag206–216, Gag241–249, Gag367–381, Vif114–124, Nef9–19, Nef89–97, and Nef193–203 epitopes, respectively. In the present study, analysis of these MHC-I haplotype 90-120-Ia-associated epitopes at 2 years showed Gag206–216, Gag241–249, and Gag367–381 epitope-specific CD8+ T-cell escape mutations in all Group M animals. Nef9–19-specific CD8+ T-cell escape mutations were also selected in most of them. However, mutations were not observed in Nef193–203 or Vif114–124 epitope-coding region. At 2 years after infection, only one of four Group M animals maintained Gag206–216/Gag241–249-specific CD8+ T cells, whereas all elicited Nef193–203 and Vif114–124 epitope-specific CD8+ T-cell responses, implying that these CD8+ T cells targeting Nef193–203 and Vif114–124 epitopes may contribute to sustained control of viremia in Group M animals.
These results suggest undetectable level of viral replication in Group M animals, resulting in accumulation of CD8+ T-cell escape mutations during viremia control. Higher SIV-specific CD8+ T-cell frequencies and broadening of the CD8+ T-cell targets in this group are considered to reflect this undetectable level of viral replication. Proviral CD8+ T-cell escape mutations were undetectable at 1 year but accumulated at 2 years in Group M, while differences in SIV non-Gag/Nef antigen-specific CD8+ T-cell responses between Groups M and N were evident as early as 4 months post-infection. This implies that broadening of the CD8+ T-cell responses in aviremic SIV controllers could serve as an indicator of the beginning of viral control failure. All the Group M animals showed reduced numbers of non-Gag/Nef antigens targeted by CD8+ T cells at 2 years compared to those at 1 year (Fig 4). It is speculated that this might be because viruses accumulate CD8+ T-cell escape mutations in those viral genome regions encoding CD8+ T cell-targeted non-Gag/Nef antigens after 1 year post-infection.
In contrast, proviral mutations did not appear in Group N macaques even at 2 years post-infection, indicating sustained viral control. These animals maintained Gag241–249-specific CD8+ T cells which are considered important for viral control. The breadth and magnitude of SIV-specific CD8+ T-cell responses remained constant in the chronic phase consistent with stable viral control in the presence of immunodominant Gag- and Nef-specific CD8+ T-cell responses. In three Group N animals (R07-001, R07-006, and R07-003), Vif-specific CD8+ T-cell responses became detectable at 2 years after infection, implying partial control failure by the Gag- and Nef-specific CD8+ T-cell responses. However, the remaining three vaccinated controllers (R07-008, R09-009, and R09-010) in Group N showed no increased breadth of their CD8+ T-cell targets, possibly reflecting the absence of escape mutants stimulating additional CD8+ T-cell responses in the chronic phase.
Effective and broad CD8+ T-cell responses have been indicated to be important for the control of HIV/SIV replication [15,47–50]. Even if aviremic HIV/SIV control is achieved by virus-specific potent CD8+ T-cell responses, residual viral replication may occur and allow accumulation of CD8+ T-cell escape mutations in viral genome, possibly leading to eventual viremia rebound. The present study addressed this issue after achievement of primary SIV control. The rhCMV vector vaccine trial indicated lasting SIV control by persistent rhCMV-induced non-classical CD8+ T-cell responses in rhesus macaques [44,51]. However, SIV controllers in the present study are thought to have no CD8+ T-cell stimulators other than virus-derived antigens post-infection. Thus, broadening of CD8+ T-cell responses is considered to be due to residual viral replication as observed in Group M animals. Broader CD8+ T-cell responses would be important for SIV control, but after achievement of a virus control condition, further broadening may indicate residual viral replication. In contrast, Group N animals, in particular three of them (R07-008, R09-009, and R09-010), showed no broadening of their CD8+ T-cell targets, which may represent a status of lasting SIV containment by CD8+ T cells.
The anti-CD8 antibody administration to the Group N macaque resulted in transient reappearance of plasma viremia concomitant with CD8+ cell depletion, supporting the notion that CD8+ T-cell responses are crucial for the stable SIV control observed. Sequence analysis of the emergent virus indicated the existence of replication-competent viruses with multiple CD8+ T-cell escape mutations, which can be controlled.
In summary, this study showed that increased breadth of virus-specific CD8+ T-cell responses is detected before the accumulation of proviral CD8+ T-cell escape mutations and viral control failure in aviremic SIV controllers. Broadly-reactive CD8+ T-cell responses may be crucial for HIV control, but our results suggest that if the host could achieve the conditions in which CD8+ T cells overwhelm HIV replication, non-broadening of CD8+ T-cell responses represents a status of lasting HIV containment by CD8+ T cells.
Materials and Methods
Ethics statement
Animal experiments were carried out in Tsukuba Primate Research Center, National Institute of Biomedical Innovation (NIBP; currently renamed National Institutes of Biomedical Innovation, Health and Nutrition [NIBIOHN]) with the help of the Corporation for Production and Research of Laboratory Primates after approval by the Committee on the Ethics of Animal Experiments of NIBP (permission number: DS21-27, DS23-19, and DS25-31) under the guideline for animal experiments at NIBP and National Institute of Infectious Diseases in accordance with the Guidelines for Proper Conduct of Animal Experiments established by Science Council of Japan (http://www.scj.go.jp/ja/info/kohyo/pdf/kohyo-20-k16-2e.pdf). The experiments were in accordance with the "Weatherall report for the use of non-human primates in research" recommendations (https://royalsociety.org/topics-policy/publications/2006/weatherall-report/)). Animals were housed in adjoining individual primate cages allowing them to make sight and sound contact with one another for social interactions, where the temperature was kept at 25°C with light for 12 hours per day. Animals were fed with apples and commercial monkey diet (Type CMK-2, Clea Japan, Inc.). Blood collection, vaccination, virus challenge, and anti-CD8 antibody treatment were performed under ketamine anesthesia.
Animal experiments
We analyzed the chronic phase of SIVmac239 infection in ten Burmese rhesus macaques (Macaca mulatta) possessing the MHC class I haplotype 90-120-Ia [32,33] (Table 1). These animals were previously used for vaccination and challenge experiments [35,36]. One-third of unvaccinated A+ animals controlled viremia after SIVmac239 infection in our previous study [35], and unvaccinated macaques R06-037 and R07-001 and sham-vaccinated R07-006 were used in the present study. Macaque R07-006 received a control prime-boost vaccine using a DNA and a replication-incompetent F-deleted SeV (F[–]SeV) vector both expressing EGFP. Three macaques R07-002, R07-003, and R07-008 received a DNA-prime/F(-)SeV-boost vaccine eliciting Gag241–249-specific CD8+ T-cell responses. A pGag236-250-EGFP-N1 DNA and an F(-)SeV-Gag236-250-EGFP vector both expressing an SIVmac239 Gag236-250 (IAGTTSSVDEQIQWM)-EGFP fusion protein were used for the single Gag241–249-epitope vaccine [35]. Three macaques R03-018, R09-009, and R09-010 received a DNA-prime/F(-)SeV-boost vaccine eliciting Gag206–216-specific CD8+ T-cell responses. A pGag202–216-EGFP-N1 DNA and an F(-)SeV-Gag202–216-EGFP vector both expressing an SIVmac239 Gag202–216 (IIRDIINEEAADWDL)-EGFP fusion protein were used for the single Gag206–216-epitope vaccine [36]. Macaque R05-005 received both the Gag241–249-epitope and Gag206–216-epitope vaccines simultaneously. Animals received 5 mg of DNA intramuscularly and 6 weeks later received a single intranasal boost with 6 x 109 cell infectious units of F(-)SeV vector. Approximately 3 months after the F(-)SeV boost, animals were challenged intravenously with 1,000 50% tissue culture infective doses of SIVmac239 [52]. For CD8+ cell depletion, macaque R09-009 received a single subcutaneous inoculation of 10 mg/kg of body weight of monoclonal anti-CD8 antibody (cM-T807) (NIH Nonhuman Primate Reagent Resource [R24 RR016001, N01 AI040101]) followed by three intravenous inoculations of 5 mg/kg cM-T807 on days 3, 7, and 10 after the first inoculation at week 108.
Proviral cDNA sequencing
Primary CD4+ T cells were prepared by negative selection from macaque PBMCs using a non-human primate CD4+ T cell isolation kit (Miltenyi). Total cellular DNA was extracted from CD4+ T cells using DNeasy extraction kit (QIAGEN). The DNA corresponding to the number of CD4+ T cells indicated in S1 Table was subjected to nested PCR amplification of proviral gag, vif, and nef cDNA fragments (nucleotide numbers [nt] 1231–2958 for gag, nt 4829–7000 for vif, and nt 8677–10196 for nef in SIVmac239 [accession number M33263]) for direct sequencing using dye terminator chemistry and an automated DNA sequencer (Applied Biosystems) as previously described [25]. For macaques R06-037, R05-005, R07-001, and R07-006 at 2 years, total cellular DNAs were extracted not directly from CD4+ T cells but after 8 days of culture described below. Dominant non-synonymous mutations were determined. For virus recovery, 0.5–2 x 106 CD4+ T cells were cultured in the presence of 10 ng/ml human interleukin-7 (IL-7) (Miltenyi) and 10 ng/ml human IL-15 (Miltenyi) for 8 days. Then, viral RNA was extracted from supernatants of CD4+ T-cell culture using the High Pure Viral RNA kit (Roche Diagnostics) and subjected to reverse transcription and nested PCR (RT-PCR) amplification of viral gag cDNA fragments.
Analysis of antigen-specific CD8+ T-cell responses
We measured virus-specific CD8+ T-cell frequencies by flow cytometric analysis of gamma interferon (IFN-γ) induction after specific stimulation as described previously [53]. Autologous herpesvirus papio-immortalized B-lymphoblastoid cell lines (B-LCLs) were pulsed with individual SIVmac239 epitope-coding peptides (at a final concentration of 1–5 μM) or peptide pools (at a final concentration of 1–2 μM for each peptide) using panels of overlapping peptides spanning the entire SIVmac239 Gag, Pol, Vif, Vpx, Vpr, Tat, Rev, Env, and Nef amino acid sequences (Sigma-Aldrich Japan) for 1 hour. PBMCs were cocultured with these pulsed B-LCLs in the presence of GolgiStop (monensin, BD) for 6 hours. Intracellular IFN-γ staining was performed with a CytofixCytoperm kit (BD) and fluorescein isothiocyanate (FITC)-conjugated anti-human CD4 (BD), peridinin chlorophyll protein (PerCP)-conjugated anti-human CD8 (BD), allophycocyanin-Cy7 (APC-Cy7)-conjugated anti-human CD3 (BD), and phycoerythrin (PE)-conjugated anti-human IFN-γ monoclonal antibodies (Biolegend). In the flow cytometric analysis, PBMCs were gated in Forward Scatter-Side Scatter dot plots and B-LCLs were excluded. A representative gating schema for flow cytometric analysis is shown in S2 Fig. Specific T-cell frequencies were calculated by subtracting nonspecific IFN-γ+ T-cell frequencies from those after peptide-specific stimulation. Specific T-cell frequencies lower than 100 per million PBMCs were considered negative.
Statistical analysis
Statistical analyses were performed with Prism software version 4.03 with significance levels set at a P value of <0.0500 (GraphPad Software, Inc.). Comparisons between groups were made using non-parametric tests.
Accession numbers
SIVmac239 proviral DNA: M33263.
Supporting Information
(TIF)
(TIF)
(PDF)
Acknowledgments
We thank F. Ono, K. Oto, A. Hiyaoka, K. Hanari, S. Okabayashi, H. Shibata, and Y. Yasutomi for their assistance in animal experiments. We also thank M. de Souza for his English proofreading and comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology in Japan (MEXT: http://www.mext.go.jp/english/) ([JSPS] KAKENHI Grants 23390115, 25893294, 15K19119, 15H04737, and 15H01271), grants-in-aid from the Ministry of Health, Labor and Welfare in Japan (MHLW: http://www.mhlw.go.jp/english/index.html) (H24-AIDS-006), and Research Programs on HIV/AIDS, Emerging and Re-emerging Infectious Diseases, and Global Health Issues from Japan Agency for Medical Research and Development (AMED: http://www.amed.go.jp/en/) (15fk0410003h0003, 15fk0410012h0001). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Rosenberg E.S., Altfeld M., Poon S.H., Phillips M.N., Wilkes B.M., Eldridge R.L., Robbins G.K., D'Aquila R.T., Goulder P.J., and Walker B.D. (2000). Immune control of HIV-1 after early treatment of acute infection. Nature 407, 523–526. [DOI] [PubMed] [Google Scholar]
- 2. Ortiz G.M., Wellons M., Brancato J., Vo H.T., Zinn R.L., Clarkson D.E., Van Loon K., Bonhoeffer S., Miralles G.D., Montefiori D., et al. (2001). Structured antiretroviral treatment interruptions in chronically HIV-1-infected subjects. Proc. Natl. Acad. Sci. U. S. A. 98, 13288–13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hocqueloux L., Prazuck T., Avettand-Fenoel V., Lafeuillade A., Cardon B., Viard J.P., and Rouzioux C. (2010). Long-term immunovirologic control following antiretroviral therapy interruption in patients treated at the time of primary HIV-1 infection. AIDS 24, 1598–1601. [DOI] [PubMed] [Google Scholar]
- 4. Saez-Cirion A., Bacchus C., Hocqueloux L., Avettand-Fenoel V., Girault I., Lecuroux C., Potard V., Versmisse P., Melard A., Prazuck T., et al. (2013). Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 9, e1003211 10.1371/journal.ppat.1003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roederer M., Keele B.F., Schmidt S.D., Mason R.D., Welles H.C., Fischer W., Labranche C., Foulds K.E., Louder M.K., Yang Z.Y., et al. (2014). Immunological and virological mechanisms of vaccine-mediated protection against SIV and HIV. Nature 505, 502–508. 10.1038/nature12893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barouch D.H., Whitney J.B., Moldt B., Klein F., Oliveira T.Y., Liu J., Stephenson K.E., Chang H.W., Shekhar K., Gupta S., et al. (2013). Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature 503, 224–228. 10.1038/nature12744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koup R.A., Safrit J.T., Cao Y., Andrews C.A., McLeod G., Borkowsky W., Farthing C., and Ho D.D. (1994). Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650–4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Borrow P., Lewicki H., Hahn B.H., Shaw G.M., and Oldstone M.B. (1994). Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68, 6103–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matano T., Shibata R., Siemon C., Connors M., Lane H.C., and Martin M.A. (1998). Administration of an anti-CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 72, 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin X., Bauer D.E., Tuttleton S.E., Lewin S., Gettie A., Blanchard J., Irwin C.E., Safrit J.T., Mittler J., Weinberger L., et al. (1999). Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189, 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schmitz J.E., Kuroda M.J., Santra S., Sasseville V.G., Simon M.A., Lifton M.A., Racz P., Tenner-Racz K., Dalesandro M., Scallon B.J., et al. (1999). Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283, 857–860. [DOI] [PubMed] [Google Scholar]
- 12. Migueles S.A., Sabbaghian M.S., Shupert W.L., Bettinotti M.P., Marincola F.M., Martino L., Hallahan C.W., Selig S.M., Schwartz D., Sullivan J., et al. (2000). HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. U. S. A. 97, 2709–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang J., Tang S., Lobashevsky E., Myracle A.D., Fideli U., Aldrovandi G., Allen S., Musonda R., Kaslow R.A., and Zambia, U.A.B.H.I.V.R.P. (2002). Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76, 8276–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kiepiela P., Leslie A.J., Honeyborne I., Ramduth D., Thobakgale C., Chetty S., Rathnavalu P., Moore C., Pfafferott K.J., Hilton L., et al. (2004). Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432, 769–775. [DOI] [PubMed] [Google Scholar]
- 15. Goulder P.J., and Watkins D.I. (2008). Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat. Rev. Immunol. 8, 619–630. 10.1038/nri2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bontrop R.E., and Watkins D.I. (2005). MHC polymorphism: AIDS susceptibility in non-human primates. Trends. Immunol. 26, 227–233. [DOI] [PubMed] [Google Scholar]
- 17. Yant L.J., Friedrich T.C., Johnson R.C., May G.E., Maness N.J., Enz A.M., Lifson J.D., O'Connor D.H., Carrington M., and Watkins D.I. (2006). The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J. Virol. 80, 5074–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loffredo J.T., Maxwell J., Qi Y., Glidden C.E., Borchardt G.J., Soma T., Bean A.T., Beal D.R., Wilson N.A., Rehrauer W.M., et al. (2007). Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J. Virol. 81, 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altfeld M., Addo M.M., Rosenberg E.S., Hecht F.M., Lee P.K., Vogel M., Yu X.G., Draenert R., Johnston M.N., Strick D., et al. (2003). Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 17, 2581–2591. [DOI] [PubMed] [Google Scholar]
- 20. Altfeld M., Kalife E.T., Qi Y., Streeck H., Lichterfeld M., Johnston M.N., Burgett N., Swartz M.E., Yang A., Alter G., et al. (2006). HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 3, e403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schneidewind A., Brockman M.A., Yang R., Adam R.I., Li B., Le Gall S., Rinaldo C.R., Craggs S.L., Allgaier R.L., Power K.A., et al. (2007). Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81, 12382–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goulder P.J., Phillips R.E., Colbert R.A., McAdam S., Ogg G., Nowak M.A., Giangrande P., Luzzi G., Morgan B., Edwards A., et al. (1997). Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3, 212–217. [DOI] [PubMed] [Google Scholar]
- 23. Feeney M.E., Tang Y., Roosevelt K.A., Leslie A.J., McIntosh K., Karthas N., Walker B.D., and Goulder P.J. (2004). Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78, 8927–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goulder P.J., and Watkins D.I. (2004). HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4, 630–640. [DOI] [PubMed] [Google Scholar]
- 25. Kawada M., Igarashi H., Takeda A., Tsukamoto T., Yamamoto H., Dohki S., Takiguchi M., and Matano T. (2006). Involvement of multiple epitope-specific cytotoxic T-lymphocyte responses in vaccine-based control of simian immunodeficiency virus replication in rhesus macaques. J. Virol. 80, 1949–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matano T., Kobayashi M., Igarashi H., Takeda A., Nakamura H., Kano M., Sugimoto C., Mori K., Iida A., Hirata T., et al. (2004). Cytotoxic T lymphocyte-based control of simian immunodeficiency virus replication in a preclinical AIDS vaccine trial. J. Exp. Med. 199, 1709–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu J., O'Brien K.L., Lynch D.M., Simmons N.L., La Porte A., Riggs A.M., Abbink P., Coffey R.T., Grandpre L.E., Seaman M.S., et al. (2009). Immune control of an SIV challenge by a T-cell-based vaccine in rhesus monkeys. Nature 457, 87–91. 10.1038/nature07469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansen S.G., Ford J.C., Lewis M.S., Ventura A.B., Hughes C.M., Coyne-Johnson L., Whizin N., Oswald K., Shoemaker R., Swanson T., et al. (2011). Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527. 10.1038/nature10003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mudd P.A., Martins M.A., Ericsen A.J., Tully D.C., Power K.A., Bean A.T., Piaskowski S.M., Duan L., Seese A., Gladden A.D., et al. (2012). Vaccine-induced CD8+ T cells control AIDS virus replication. Nature 491, 129–133. 10.1038/nature11443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matano T., Kano M., Nakamura H., Takeda A., and Nagai Y. (2001). Rapid appearance of secondary immune responses and protection from acute CD4 depletion after a highly pathogenic immunodeficiency virus challenge in macaques vaccinated with a DNA prime/Sendai virus vector boost regimen. J. Virol. 75, 11891–11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kawada M., Tsukamoto T., Yamamoto H., Iwamoto N., Kurihara K., Takeda A., Moriya C., Takeuchi H., Akari H., and Matano T. (2008). Gag-specific cytotoxic T-lymphocyte-based control of primary simian immunodeficiency virus replication in a vaccine trial. J. Virol. 82, 10199–10206. 10.1128/JVI.01103-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Naruse T.K., Chen Z., Yanagida R., Yamashita T., Saito Y., Mori K., Akari H., Yasutomi Y., Miyazawa M., Matano T., et al. (2010). Diversity of MHC class I genes in Burmese-origin rhesus macaques. Immunogenetics 62, 601–611. 10.1007/s00251-010-0462-z [DOI] [PubMed] [Google Scholar]
- 33. Nomura T., Yamamoto H., Shiino T., Takahashi N., Nakane T., Iwamoto N., Ishii H., Tsukamoto T., Kawada M., Matsuoka S., et al. (2012). Association of major histocompatibility complex class I haplotypes with disease progression after simian immunodeficiency virus challenge in burmese rhesus macaques. J. Virol. 86, 6481–6490. 10.1128/JVI.07077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nomura T., Yamamoto H., Takahashi N., Naruse T.K., Kimura A., and Matano T. (2014). Identification of SIV Nef CD8(+) T cell epitopes restricted by a MHC class I haplotype associated with lower viral loads in a macaque AIDS model. Biochem. Biophys. Res. Commun. 450, 942–947. 10.1016/j.bbrc.2014.06.072 [DOI] [PubMed] [Google Scholar]
- 35. Tsukamoto T., Takeda A., Yamamoto T., Yamamoto H., Kawada M., and Matano T. (2009). Impact of cytotoxic-T-lymphocyte memory induction without virus-specific CD4+ T-Cell help on control of a simian immunodeficiency virus challenge in rhesus macaques. J. Virol. 83, 9339–9346. 10.1128/JVI.01120-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ishii H., Kawada M., Tsukamoto T., Yamamoto H., Matsuoka S., Shiino T., Takeda A., Inoue M., Iida A., Hara H., et al. (2012). Impact of vaccination on cytotoxic T lymphocyte immunodominance and cooperation against simian immunodeficiency virus replication in rhesus macaques. J. Virol. 86, 738–745. 10.1128/JVI.06226-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moriya C., Igarashi H., Takeda A., Tsukamoto T., Kawada M., Yamamoto H., Inoue M., Iida A., Shu T., Hasegawa M., et al. (2008). Abrogation of AIDS vaccine-induced cytotoxic T-lymphocyte efficacy in vivo due to a change in viral epitope flanking sequences. Microbes Infect. 10, 285–292. 10.1016/j.micinf.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 38. Sheehy A.M., Gaddis N.C., Choi J.D., and Malim M.H. (2002). Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650. [DOI] [PubMed] [Google Scholar]
- 39. Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., and Malim M.H. (2003). DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809. [DOI] [PubMed] [Google Scholar]
- 40. Zennou V., and Bieniasz P.D. (2006). Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology 349, 31–40. [DOI] [PubMed] [Google Scholar]
- 41. Fourati S., Lambert-Niclot S., Soulie C., Malet I., Valantin M.A., Descours B., Ait-Arkoub Z., Mory B., Carcelain G., Katlama C., et al. (2012). HIV-1 genome is often defective in PBMCs and rectal tissues after long-term HAART as a result of APOBEC3 editing and correlates with the size of reservoirs. J. Antimicrob. Chemother. 67, 2323–2326. 10.1093/jac/dks219 [DOI] [PubMed] [Google Scholar]
- 42. Eisele E., and Siliciano R.F. (2012). Redefining the viral reservoirs that prevent HIV-1 eradication. Immunity 37, 377–388. 10.1016/j.immuni.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Whitney J.B., Hill A.L., Sanisetty S., Penaloza-MacMaster P., Liu J., Shetty M., Parenteau L., Cabral C., Shields J., Blackmore S., et al. (2014). Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 512, 74–77. 10.1038/nature13594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hansen S.G., Piatak M. Jr., Ventura A.B., Hughes C.M., Gilbride R.M., Ford J.C., Oswald K., Shoemaker R., Li Y., Lewis M.S., et al. (2013). Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104. 10.1038/nature12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Eriksson S., Graf E.H., Dahl V., Strain M.C., Yukl S.A., Lysenko E.S., Bosch R.J., Lai J., Chioma S., Emad F., et al. (2013). Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 9, e1003174 10.1371/journal.ppat.1003174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ho Y.C., Shan L., Hosmane N.N., Wang J., Laskey S.B., Rosenbloom D.I., Lai J., Blankson J.N., Siliciano J.D., and Siliciano R.F. (2013). Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551. 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ndhlovu Z.M., Proudfoot J., Cesa K., Alvino D.M., McMullen A., Vine S., Stampouloglou E., Piechocka-Trocha A., Walker B.D., Pereyra F. (2012). Elite controllers with low to absent effector CD8+ T cell responses maintain highly functional, broadly directed central memory responses. J. Virol. 86, 6959–6969. 10.1128/JVI.00531-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Budde M.L., Greene J.M., Chin E.N., Ericsen A.J., Scarlotta M., Cain B.T., Pham N.H., Becker E.A., Harris M., Weinfurter J.T., et al. (2012). Specific CD8+ T cell responses correlate with control of simian immunodeficiency virus replication in Mauritian cynomolgus macaques. J. Virol. 86, 7596–7604 10.1128/JVI.00716-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Radebe M., Gounder K., Mokgoro M., Ndhlovu Z.M., Mncube Z., Mkhize L., van der Stok M., Jaggernath M., Walker B.D., Ndung'u T. (2015). Broad and persistent Gag-specific CD8+ T-cell responses are associated with viral control but rarely drive viral escape during primary HIV-1 infection. AIDS 29, 23–33. 10.1097/QAD.0000000000000508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deng K., Pertea M., Rongvaux A., Wang L., Durand C.M., Ghiaur G., Lai J., McHugh H.L., Hao H., Zhang H., et al. (2015). Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385. 10.1038/nature14053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hansen S.G., Sacha J.B., Hughes C.M., Ford J.C., Burwitz B.J., Scholz I., Gilbride R.M., Lewis M.S., Gilliam A.N., Ventura A.B., et al. (2013). Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 10.1126/science.1237874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kestler H.W. 3rd, Ringler D.J., Mori K., Panicali D.L., Sehgal P.K., Daniel M.D., and Desrosiers R.C. (1991). Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65, 651–662. [DOI] [PubMed] [Google Scholar]
- 53. Iwamoto N., Takahashi N., Seki S., Nomura T., Yamamoto H., Inoue M., Shu T., Naruse T.K., Kimura A., and Matano T. (2014). Control of simian immunodeficiency virus replication by vaccine-induced Gag- and Vif-specific CD8+ T cells. J. Virol. 88, 425–433. 10.1128/JVI.02634-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.