Fig. 3.
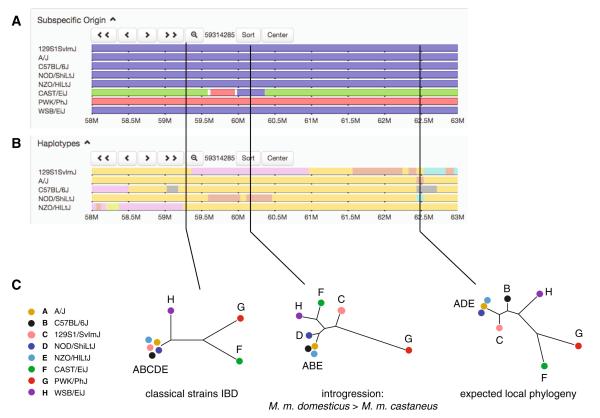
Exploring local haplotype diversity in the Collaborative Cross founder strains. a Subspecific origin tracks from the Mouse Phylogeny Viewer for a five Mbp interval on chromosome 2. Segments are colored according to the subspecies from which they were most likely inherited: blue for M. m. domesticus, red for M. m. musculus, and green for M. m. castaneus. As expected, the five classical laboratory strains are of mostly M. m. domesticus ancestry, but an introgression tract (see Appendix), from M. m. musculus into the classical strain NZO/HILtJ, is visible in the distal portion of the interval. b Fine-scale haplotype block maps for the five classical laboratory strains. (The three wild-derived founder strains are excluded from this analysis because each has a private haplotype, shared with none of the other founder strains, in almost every genomic interval. See Appendix.) Strains with the same haplotype at a given position are assigned the same color, but colors are recycled along the length of the window. c Local phylogenetic trees reflect the varying ancestry of the CC founder strains along the genome. From left: an interval in which all five classical strains share a haplotype identical-by-descent (IBD, see Appendix); an interval in which CAST/EiJ clusters within a class of classical inbred strains, reflecting introgression (probably due to breeding errors in the laboratory); and an interval whose phylogeny is consistent with the genome-wide expected relationship between strains (see Fig. 1).In the absence of epistasis, allele effects at a QTL should be concordant with the local phylogenetic tree: for instance, in the middle interval, the effect of the PWK/PhJ allele should differ from that of any of the other seven alleles, and the effects of the other seven should be similar to each other