Fig. 3.
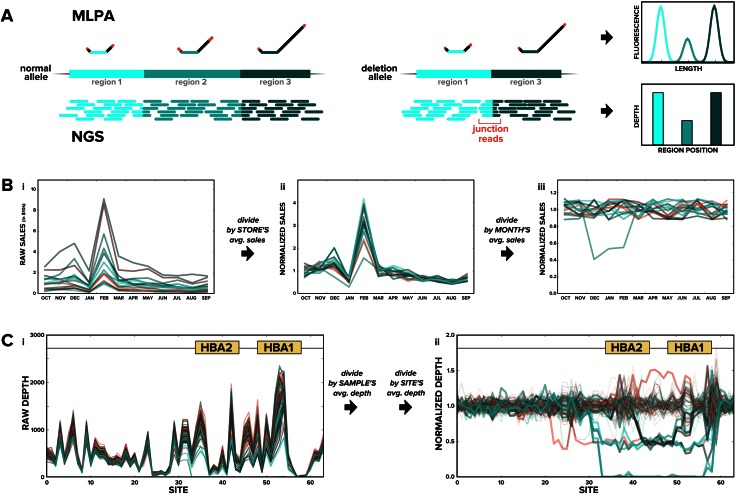
Del/dup calling from NGS data requires simple and intuitive processing of raw data. a Schematic of MLPA (top) and NGS (bottom) data for a sample in which one chromosome is normal and the other has a deletion. MLPA probes have a genome-binding sequence (shades of green), stuffer sequence to give them unique length (black), and binding sites for common primers (red) at the termini that enable multiplex amplification. For NGS, read depth can be pooled across a region (as depicted) or counted at a single site. In addition to depth data supporting a del/dup, NGS provides evidence of junction reads that further support the observation of a del/dup. For clarity, the ligation step that fuses two DNA fragments into the probes depicted in the figure is omitted. b A chocolate store that underperforms relative to others is revealed by dividing (i) the hypothetical annual sales volume for each store by its average (yielding ii) and then dividing once more by the monthly average across stores (giving iii). c Multiple samples with del/dups in the HBA locus are discovered by normalizing (i) the raw depth data across many sites by the sample average and then by the site average (yielding ii, where del/dup samples have thick traces)