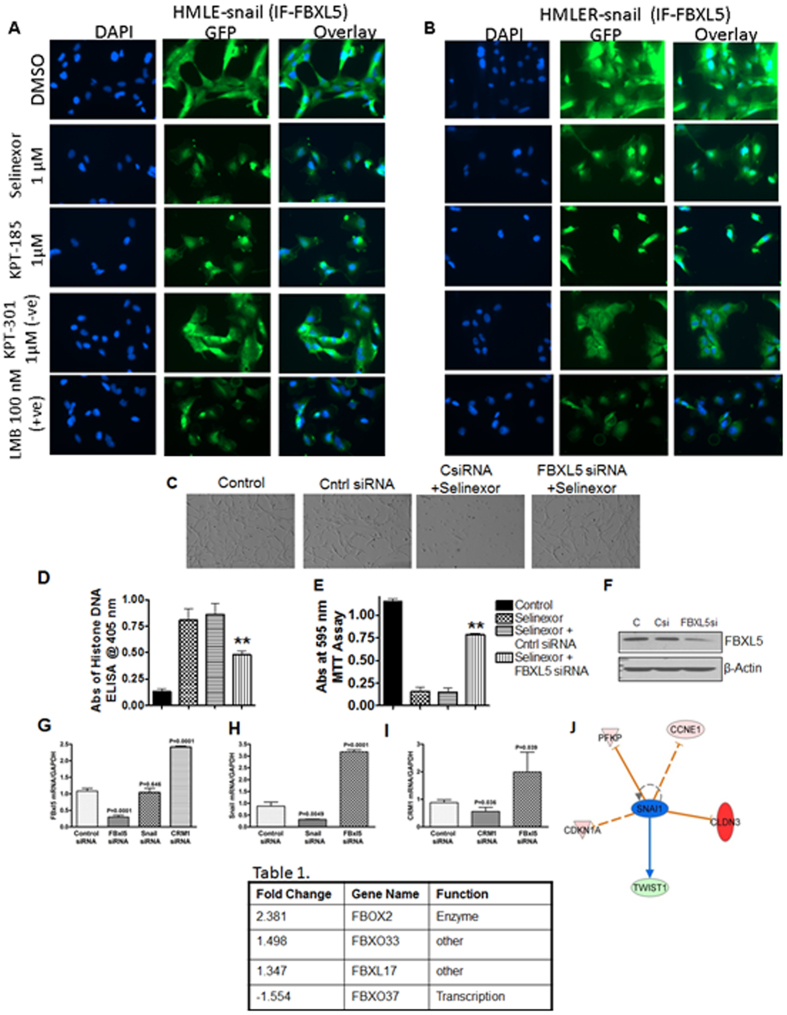
Figure 4. Role of FBXL5 in snail turnover and reversal of EMT.
(A,B) Cells growing in chambered slides (3000 cells per well) overnight and exposed to SINE compounds at indicated concentrations for an additional 24 hrs. Immunofluorescence assay was performed as described in methods section. Immunofluorescence images (40×) of HMLE-snail and HMLER-snail cells showing nuclear retention of FBXL5 (Abcam, USA). (C) Photomicrographs of HMLE-snail cells under different treatment conditions. Note. Lack of EMT reversal in the presence of FBXL5 siRNA. (D) Histone DNA ELISA apoptosis and (E) MTT assay at 72 hrs post Selinexor treatment (150 nM) in the absence or presence of siRNAs for FBXL5 (see methods section for siRNA procedures). **P < 0.01 (siRNA vs untreated groups). (F) Western blot showing siRNA silencing results in suppression of total FBXL5. (G–I) RT-PCR analysis post FBXL5 siRNA, snail siRNA or CRM1 siRNA silencing respectively. HMLE-snail cells were exposed to FBXL5 siRNA according to procedure described in Methods section. RNA was isolated and RT-PCR was performed. The relative expression of all mRNAs tested by qRT-PCR in control siRNA treated cells were normalized to 1.0 (unit value) and were compared with their respective siRNA treated cells. (J) Computational analysis of SINE induced reversal of Mesenchymal phenotype. HMLE-snail cells were seeded in quadruplets in 100 mm petri dishes overnight. Cells were exposed to KPT-185 (150 nM) for additional 24 hrs followed by gene expression microarray analysis (Agilent platform). To construct a p-value for each gene, the test statistics from the 4 replicate experiments were used in a 3 degree of freedom t-test analysis. This t-test is testing whether the average test statistics is not equal to 0 (0 would be no difference treated v. untreated). Each gene thus has a mean log ratio value and a p-value for the detected ratio which determines the significance of the mean log ratio (treated/untreated expression levels combining all 4 replicates). Our objective was to discover what was changed repeatedly in all 4 biological replicates. Statistically significant (p = 0.001) and differentially expressed genes were subjected to Ingenuity Pathway Analysis (IPA) which showed down regulation of Snail network. [Table 1] Some of the F-BOX genes that were differentially expressed (p < 0.001) are tabulated. See Supplementary Table 1 for detailed information on the list of genes differentially expressed.