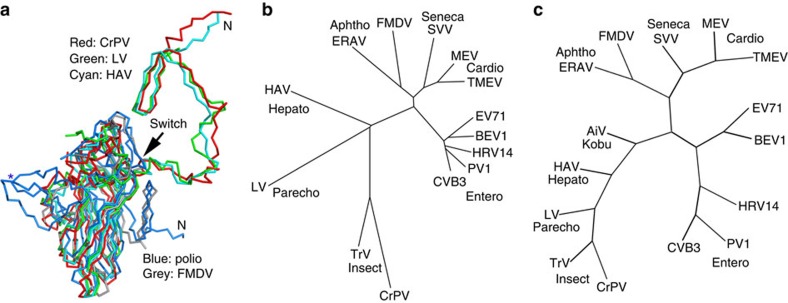
Figure 5. Phylogeny.
(a) Superposition of LV VP0 with VP2 of poliovirus, FMDV, HAV and CrPV. The N terminus of LV VP0 folds similarly to that of HAV and CrPV but differently to other picornaviruses. The star marks the EF loop that forms part of the south wall of the canyon in poliovirus. (b) Structure and (c) sequence-based phylogenetic trees of representative picornaviruses and cripaviruses, respectively: CVB3, coxsackievirus B3; PV1, poliovirus type 1; HRV14, human rhinovirus 14; BEV1, bovine enterovirus type 1; EV71, EV-A71; TMEV, Theilers virus; MEV, Mengo virus; SVV, Seneca valley virus; FMDV, foot-and-mouth disease virus; ERAV, equine rhinitis A virus; HAV, hepatitis A virus; AiV, Aichi virus59; TrV, triatoma virus; CrPV, cricket paralysis virus.