Abstract
Clear cell renal cell carcinoma (ccRCC), the most aggressive and lethal form of renal cell carcinoma accounts for over 90% of metastasis that occur following curative surgery for clinically confined disease. High relapse rates have prompted the evaluation of targeted therapies for the prevention or delay of metastatic disease in high-risk patients, with biomarkers offering significant potential to guide and improve patient management in this setting. In this current study we examined the value of the 4E-BP1/eIF4E axis for prognostic significance and risk stratification in patients with clinically confined ccRCC. This axis is a critical convergence point for many signalling pathways that are targeted by current therapies for the treatment of advanced RCC. Immunohistochemistry for phosphorylated 4E-BP1 (p4E-BP1) and total eIF4E was performed on tissue microarrays containing tumour cores from 135 patients with localised ccRCC. For both biomarkers 39% of all evaluable cores stained positive, with a strong correlation observed between the presence of p4E-BP1 and the overexpression of eIF4E within the same tumour (P = 0.005). Further, the combined expression of p4E-BP1 and eIF4E was associated with significantly worse disease-free survival of 2.9 vs 5.7 yrs compared to patients whose tumours expressed only one, or neither, of the biomarkers (P < 0.001). Cox-regression analysis confirmed the ability of the p4EBP1/eIF4E signature to independently identify high-risk patients with a Hazard Ratio of 4.2 (CI = 2.1-8.6; P < 0.001), compared to 3.3 for tumour grade 3 and 4, and 2.3 for tumour stage 3 and 4. These data show the powerful prognostic value of the p4E-BP1/eIF4E signature for potential management of patients with clinically confined ccRCC, and in addition provides insights into the possible key synergistic determinants of disease progression and treatment response.
Keywords: Clear cell renal cell carcinoma, ccRCC, metastatic disease, 4E-BP1, eIF4E, biomarker, prognosis, stratification, immunohistochemistry, mTOR
Introduction
Renal cell carcinoma (RCC) is a heterogeneous and complex family of kidney tumours that are comprised of several distinct subtypes of which the clear cell subtype (ccRCC) comprises 70-80% of cases and more than 90% of RCC tumour deaths [1]. The incidence of RCC appears to be rising steadily at a rate of 2.5% per year [2]. These increases reflect the detection of incidental tumours through improved abdominal imaging [3] and the increased prevalence of risk factors such as smoking, obesity and hypertension [2]. The majority of sporadic ccRCC’s arise from loss of function mutations or biallelic hypermethylation of the VHL gene [4]. At first presentation one-third of all patients with RCC will have established metastatic tumours (mRCC) and despite the introduction of molecular targeted therapies the overall 5 year survival rate of this patient group rarely exceeds 10%. Patients who present with clinically confined disease will usually undergo curative nephrectomy, however, up to 40% will eventually relapse with secondary tumours at distant sites. The clinical course in localised ccRCC is difficult to predict, even within patients who have similar clinico-pathological parameters ie. tumour stage and grade, presence of vascular and capsular invasion.
The identification of molecular signatures that accurately reflect biologically relevant pathways in RCC should prove valuable for predicting the behaviour of tumours and their response to treatment [5]. The incorporation of predictive tumour biomarkers in prognostication schemes for surveillance purposes and treatment choice is increasingly implemented [5,6]. The number of potential tumour biomarkers reported for RCC has increased exponentially over the last decade but few have proven to have valid clinical utility. Reliable biomarkers which provide prognostic or predictive evidence and add further value above and beyond conventional histological parameters are still needed to guide patient management [7].
In recent years, much interest has focussed on the mTOR signalling pathway as a suitable drug target for the treatment of advanced cancers, particularly for mRCC. Distinct signalling cascades such as the EGFR/ERK, PI3-K/AKT and IGFR pathways are known to converge at mTOR to modulate cell growth, migration and invasion. The two most studied immediate effector molecules of mTOR are the 4E-binding protein 1 (4E-BP1) and p70SK6, which together comprise the TORC1 component of the mTOR signalling hub. The mTOR mediated phosphorylation of p70SK6 activates the 40S ribosomal S6 kinase at serine-235 and serine-236 thereby initiating ribosomal biogenesis. Meanwhile, inhibitory phosphorylation of 4E-BP1 at serine-65 represents an important terminal step of a hierarchal phosphorylation cascade that reduces the ability of 4E-BP1 to bind and inactivate the translation-initiation factor eIF4E. Clinical studies have shown the inactivated phosphorylated form of 4E-BP1 (p4E-BP1) to be the most accurate single biomarker of the mTOR pathway for the prediction of disease progression in carcinomas of the ovary [8], brain [9] and prostate [10].
At the normal low cellular levels the eIF4E molecule is a rate-limiting factor governing the kinetics of protein synthesis [11]. Under high levels of expression such as in cancer cells, eIF4E favours the translation of mRNA subsets that have complex and highly structured 5-termini [12]. These proteins often have short-half lives and include the oncogenic growth promoting proteins cyclin D1 and c-myc and the anti-apoptotic molecules survivin and Mcl-1 [11]. Functional studies have shown eIF4E to be a bone fide oncogene where its forced overexpression is sufficient alone to cause cell immortalisation and abrogation of apoptosis leading to chemo- and radio-resistance in cancer cells [13]. For example, in mice the overexpression of eIF4E is able to sustain the growth of human lymphoma xenographs [14]. While in cancer cell lines derived from the prostate [10], breast [15] and endometrium [16] the targeted downregulation of eIF4E is shown to induce cell death and suppress cell growth and invasion. Consistent with these pro-oncogenic properties elevated levels of total eIF4E are reported in many advanced cancers including malignancies of the oesphagus [17], breast [18], liver [19] and lung [20]. Phosphorylation of eIF4E at residue serine-209 by MAPK-interacting proteins, MNK1 and MNK2, leads to increases in protein synthesis and promotes tumorigenesis. However, the exact role of phosphorylated eIF4E in oncogenic transformation and cancer maintenance remains uncertain as tumour levels of phosphorylated eIF4E do not appear to correlate with disease progression or other aggressive features [11,21].
This current study was conducted with archival primary tumour material from a consecutive series of 135 patients diagnosed with ccRCC and who had previously undergone presumptive curative surgery for localised disease. We found that the presence of p4E-BP1 in the primary tumour correlates in a highly significant manner with the over-expression of eIF4E, and that both p4E-BP1 and eIF4E serve as independent biomarkers for disease recurrence. Moreover this biomarker combination identifies a subset of patients with aggressive disease who exhibit significantly poorer disease free survival. Multivariate analysis showed that the composite co-variate of p4E-BP1/eIF4E to be a powerful predictor of early relapse with a hazard ratio of 4.2 (vs 2.3 for stage pT3 & pT4 and 3.3 for Grade 3 & 4 disease). We propose that p4E-BP1 and eIF4E together represent a biologically relevant molecular signature in ccRCC for the prediction of disease progression and clinical trial patient stratification which seeks to evaluate cognate molecular targeted therapies.
Materials and methods
Antibodies and reagents
Rabbit polyclonal antihuman antibodies for p4E-BP1 (Ser65) and total eIF4E were purchased from New England Biolabs (Herts, UK). Swine anti-rabbit secondary HRP-conjugated antibodies and non-immune rabbit serum were obtained from DAKO (Cambridge, UK). All antibodies and reagents were used according to the manufacturer’s instructions.
Patient selection and tissue microarray construction
The patient cohort, previously described in part elsewhere [22-24], consisted of a consecutive series of 175 patients, who had undergone curative radical nephrectomy between 1992 and 1999 for primary RCC. Paraffin blocks, histology reports and slides were available in all cases. Sections were reexamined and the tumours re-staged to UICC 2002 and tumour type classified according to the WHO 2004 classification; all tumours classified as clear cell renal cell carcinoma (ccRCC) were selected for study (135 cases). Also recorded were; Fuhrman nuclear grade [25], the presence or absence of any vascular invasion (either microvascular invasion, renal vein invasion or inferior vena cava invasion) [22], and whether or not there was capsular invasion with cellular invasion of peri-nephric or renal sinus fat [26]. The ECOG-Performance status of the patients was not available. None had received treatment or had evidence of lymph node or distant metastatic disease prior to or at surgery.
Clinical follow-up was carried out as previously described [22,23]. Patients had usually been reviewed annually as an out-patient for between 3-8 years; the following information was extracted from the patient records: date of birth, gender, date of surgery, date last seen, date of death, cause of death and the date on which recurrent or metastatic disease was first identified.
For each renal carcinoma a paraffin-embedded block was selected that contained a sample of peripheral tumour that could be used in the tissue microarray (TMA). A single core of representative peripheral tumour, 0.6 mm in diameter, was punched from each donor block and using a specific orientation transplanted into a pre-moulded recipient paraffin wax block. Additional cores were taken from normal renal tissue (adjacent to some of the tumours) and from human placenta. Serial sections were cut at 4 µm thickness from the resulting TMA block and laid onto clean adhesive glass slides (Superfrost PlusTM).
Immunocytochemistry and staining interpretation
TMA sections were deparaffinised and rehydrated using standard methodologies as previously described [23]. Endogenous peroxidase activity within the rehydrated tissue was inactivated with 3% hydrogen peroxide in methanol for 10 min at room temperature. Antigen retrieval was carried out by boiling in sodium citrate solution (pH 6.0) for 10 min and then cooled and equilibrated in OptimaxTM wash buffer. The TMA sections were incubated (15 hr at 4°C) with the primary antibodies for eIF4E (dilution 1:25), and p4E-BP1 (dilution 1:25) with the diluent 0.6% BSA in OptimaxTM wash buffer. After washing (4 × 1 min), sections were incubated with the appropriate secondary HRP-conjugated antibody at a dilution of 1:100 for 1 hr at room temperature. After washing the sections were developed using 3,3’-diaminobenzidine (Sigma, Poole, UK) and then counterstained with haemotoxylin, dehydrated and mounted. Negative controls consisted of sections where the primary antibody had been omitted or replaced with non-immune rabbit serum. Positive controls for p4E-BP1 and eIF4E immunoreactivity consisted of human placenta tissue cores incorporated into the TMA.
Scoring of TMA sections were undertaken by a pathologist (DFRG) and research associate (LC) using a double-headed light microscope without prior knowledge of clinical outcome as previously described [23,24]. Expression of each marker was assessed semi-quantitatively according to a previously validated criteria that accounted for both the intensity of immuno-stain and percentage of tumour cells involved within each core. Scoring was as follows: 0: no detectable deposit in tumour cells; 1: very light diffuse or focal light deposit in tumour cell cytoplasm; 2: light diffuse or moderate focal deposit (but may include very small areas of heavy deposit); 3: tumour containing areas of heavy deposit in most or all tumour cells. These scores were then converted to a simple binary score of either negative or positive according to the most informative split by Kaplan-Meier analysis. Any staining for either eIF4E or p4E-BP1 was considered positive while a complete absence of staining was considered negative.
Statistical analysis
Correlations between biomarker expression and prognostic clinico-pathological variables (tumour stage, grade and size; presence or absence of vascular invasion and capsular invasion) was examined by crosstabulation and chi-squared testing or Fishers exact test as appropriate. Univariate analysis of disease free survival (DFS) of patients with tumours showing different scores of staining for each biomarker was carried out by the Kaplan-Meier method using the log rank statistical test. The first appearance of a metastasis was considered an event. Patients last seen alive without metastasis or who died due to other causes other than RCC were considered censored at the date last seen or date of death, respectively. Multivariate survival analysis for p4E-BP1 and eIF4E was carried out by Cox regression using the Enter or Forward Stepwise (Likelihood Ratio) function with the covariates grade and stage entered in the final model.
The statistical package SPSS 11.5 was used for analysis. All tests were two tailed.
Results
Presence of phosphorylated 4E-BP1 correlates in a highly significant manner with the overexpression of eIF4E in clinically-confined ccRCC tumours
A summary of patient details are shown in Table 1. Of the 135 cases of clinically confined ccRCC 113 cases were assessable for the measurement of p-4EBP1 stain. Of these 43 (38%) stained positively for p-4EBP1 with: 33 cases scored at 1, 9 cases scored at 2, and 1 case scored at 3. Similarly for eIF4E 129 cases were assessable for the measurement of stain with 48 (37%) cases staining positively for eIF4E with of these: 28 cases scored at 1, 18 cases scored at 2, and 2 cases scored 3. Typical cores showing positive and negative staining for p4E-BP1 and eIF4E are shown in Figure 1. The intra-tumoural co-expression of p4E-BP1 and eIF4E could be assessed in 111 cases with 21 (19%) of these tumours positively expressing both biomarkers (Table 2). Cross-tabulation of p4E-BP1 with eIF4E showed a highly significant correlation with the p4E-BP1 positive staining indicating a statistically significant (P = 0.005) increased likelihood of eIF4E over-expression. These data suggest that the phosphorylation of 4E-BP1 in ccRCC tumours is a distinct biological event that can functionally lead to the over-expression of eIF4E.
Table 1.
Summary of patient details
| Patient breakdown (= 135) | ||
|---|---|---|
| Age (years) | Median | 63 |
| Min | 33 | |
| Max | 86 | |
| Gender | Male | 89 |
| Female | 46 | |
| Grade | 1 | 18 |
| 2 | 81 | |
| 3 | 25 | |
| 4 | 11 | |
| T Stage | 1 | 52 |
| 2 | 30 | |
| 3 & 4 | 53 | |
| Size (cm) | < 4 | 24 |
| 4-7 | 49 | |
| 7-10 | 42 | |
| > 10 | 20 |
Figure 1.
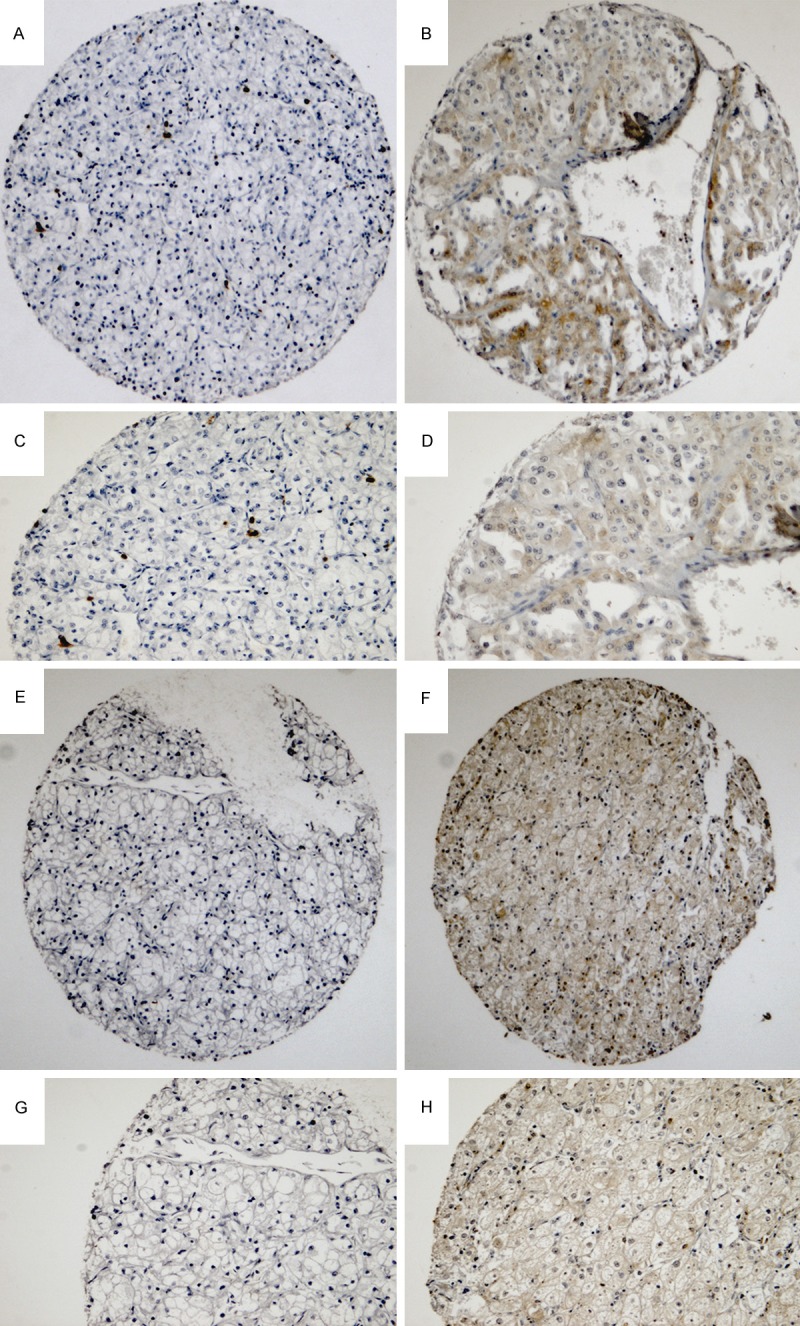
Representative cores from a TMA of clinically confined clear cell renal cell carcinoma. Images demonstrate the negative and positive immunohistochemical staining pattern for p4E-BP1 (A to D) and eIF4E (E to H). Typical cores for p4E-BP1 and eIF4E are shown in low (A, B, E, F) and high magnification (C, D, G, H).
Table 2.
Relationship of p4E-BP1, EIF4E, and combined p4E-BP1/EIF4E with clinico-pathological parameters in clinically confined ccRCC
| p4E-BP1 (n = 113) | EIF4E (n = 129) | Combined (n = 111) | ||||
|---|---|---|---|---|---|---|
| Grade | -ve | +ve | -ve | +ve | -ve | +ve |
| Grade 1 & 2 | 56 | 29 | 62 | 33 | 71 | 12 |
| Grade 3 | 11 | 8 | 13 | 10 | 14 | 5 |
| Grade 4 | 3 | 6 | 6 | 5 | 5 | 4 |
| P = 0.15 | P = 0.62 | P = 0.062 | ||||
| Size | ||||||
| 4 cm and less | 11 | 7 | 16 | 6 | 14 | 2 |
| > 4 cm to 7 cm | 24 | 16 | 27 | 20 | 31 | 9 |
| > 7 to 10 cm | 26 | 11 | 26 | 14 | 31 | 6 |
| > 10 cm | 9 | 9 | 12 | 8 | 14 | 4 |
| P = 0.52 | P = 0.65 | P = 0.78 | ||||
| Stage | ||||||
| Stage 1 | 26 | 15 | 31 | 19 | 33 | 6 |
| Stage 2 | 14 | 10 | 21 | 7 | 20 | 4 |
| Stage 3 & 4 | 30 | 18 | 29 | 22 | 37 | 11 |
| P = 0.27 | P = 0.27 | P = 0.63 | ||||
| Vascular Invasion | ||||||
| (-ve) | 40 | 23 | 48 | 25 | 53 | 8 |
| (+ve) | 30 | 20 | 33 | 23 | 37 | 13 |
| P = 1 | P = 0.42 | P = 0.08 | ||||
| Capsular Invasion | ||||||
| (-ve) | 56 | 34 | 66 | 39 | 72 | 16 |
| (+ve) | 14 | 9 | 15 | 9 | 18 | 5 |
| P = 0.9 | P = 0.98 | P = 0.70 | ||||
P < 0.05 denotes significance.
Lack of association of p4E-BP1 and eIF4E with conventional histological parameters of clinically-confined ccRCC
Cross-tabulation of p4E-BP1 and eIF4E as individual biomarkers, or as a combined co-variate, failed to demonstrate any significant association with conventional clinico-pathological parameters (Table 2). A strong trend for the combined expression of p4E-BP1 and eIF4E with high tumour grade was however observed (P = 0.062). This data suggests that the presence of one or both p4E-BP1 and eIF4E can occur as an early event in the pathogenesis of ccRCC and is not necessarily secondary to any specific aggressive histological parameter.
Tumour expression of p4E-BP1 and/or eIF4E leads to poor disease-free survival in patients with clinically-confined ccRCC
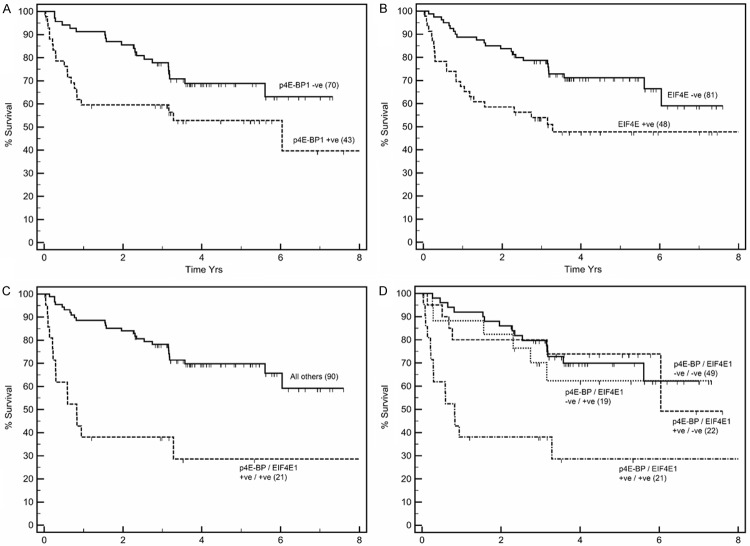
Univariate survival analysis showed that patients whose tumours stained positive for p4E-BP1 had significantly shorter time to relapse, with a mean disease-free survival (DFS) of 4.4 vs 5.6 yrs compared to patients whose tumours were negative (P = 0.02) (Figure 2A). Similarly, patients whose tumours expressed eIF4E had a mean DFS of 4.4 vs 5.7 yrs compared to patients whose tumours were negative (P = 0.005) (Figure 2B). Patients whose tumours co-expressed p4E-BP1 and eIF4E had a significantly worse mean DFS of 2.9 vs 5.7 yrs compared to patients whose tumours expressed only one, or neither, of the biomarkers (P < 0.001) (Figure 2C).
Figure 2.
Kaplan-Meier disease-free survival plots of patients presenting with clinically confined ccRCC whose primary tumours were stratified as positive (+) or negative (-) for expression of p4E-BP1 (A), eIF4E (B), co-expression of p4E-BP1 and eIF4E (C), and sub-analysis (D) of patients who were either negative for both p4E-BP1 and eIF4E; positive for p4E-BP1 alone; positive for eIF4E alone; positive for both biomarkers. Patients whose tumours expressed both p4E-BP1 and eIF4E had significantly worse prognosis.
To assess the influence of p4E-BP1 and eIF4E interaction on disease progression an analysis was carried out stratifying patients into four groups according to combination of expression of biomarkers p4E-BP1 and eIF4E (Figure 2D). This stratification revealed patients whose tumours showed no expression of both p4E-BP1 and eIF4E (double negative cases, n = 49) to have a mean DFS of 5.4 yrs. This DFS was similar to that of patients whose tumours expressed either p4E-BP1 or eIF4E alone (i.e. single positive cases n = 22, DFS 5.6, and n = 19 DFS 5.3, respectively). However, tumours that expressed both biomarkers (double positive cases) had a significantly worse (P = 0.02) DFS of 2.9 yrs. Thus the influence of each biomarker on the outcome appeared to be due to the inclusion of double positive cases in the group suggesting the phosphorylation of 4E-BP1 and the overexpression eIF4E are linked molecular events that co-operate to drive disease progression in clinically-confined ccRCC.
Cox-regression analyses was undertaken to determine the prognostic value of p4E-BP1 and eIF4E, either as single co-variates or as a composite covariate together with tumour stage and grade. We found p4E-BP1 and p4E-BP1 and eIF4E were both significant predictors of shortened DFS with (Tables 3, 4 and 5). Notably, the composite co-variate of p4E-BP1 and eIF4E was a significant and powerful predictor of disease recurrence (Table 5) with a HR of 4.2 (CI = 2.1-8.6; P < 0.001), whereas the HR for tumour grade was 3.3 (CI = 2.0-5.2; P < 0.001) and for stage III and IV was = 2.3 (CI = 2.3-3.6; P < 0.001).
Table 3.
Multivariate Cox regression hazard model for time to recurrence for p4E-BP1 as calculated by Cox proportional hazard computation including tumour grade and tumour stage
| Prognostic indice | HR | 95% CI | P value |
|---|---|---|---|
| Grade 1 and 2 | 1 | ||
| Grade 3 and 4 | 3.5 | 2.1-5.6 | < 0.001 |
| Stage I and II | 1 | ||
| Stage III and IV | 2.3 | 1.4-3.6 | < 0.001 |
| p4E-BP1 covariate negative | 1 | ||
| p4E-BP1 covariate positive | 2.3 | 1.2-4.3 | 0.01 |
P < 0.05 denotes significance.
Table 4.
Multivariate Cox regression hazard model for time to recurrence for eif4e as calculated by Cox proportional hazard computation including tumour grade, tumour stage and tumour size
| Prognostic indice | HR | 95% CI | P value |
|---|---|---|---|
| Grade 1 and 2 | 1 | ||
| Grade 3 and 4 | 3.7 | 2.4-5.6 | < 0.001 |
| Stage I and II | 1 | ||
| Stage III and IV | 2.0 | 1.4-2.9 | < 0.001 |
| eIF4E covariate negative | 1 | ||
| eIF4E covariate positive | 2.3 | 1.3-4.1 | 0.006 |
P < 0.05 denotes significance.
Table 5.
Multivariate Cox regression hazard model for time to recurrence for the composite covariate of eIF4E and p4E-BP1
| Prognostic indice | HR | 95% CI | P value |
|---|---|---|---|
| Grade 1 and 2 | 1 | ||
| Grade 3 and 4 | 3.3 | 2.0-5.2 | < 0.001 |
| Stage I and II | 1 | ||
| Stage III and IV | 2.3 | 2.3-3.6 | < 0.001 |
| eIF4E-p4E-BP1 covariate negative | 1 | ||
| eIF4E-p4E-BP1 covariate positive | 4.2 | 2.1-8.6 | < 0.001 |
Tumour size and vascular invasion were also entered into the model but were not considered significant at this step. P < 0.05 denotes significance.
Discussion
Relapse rates for clinically confined RCC are frequently reported at 25% to 40% with the ability to accurately identify patients at high-risk of metastatic disease remaining a major challenge. Conventional clinico-pathological parameters appear not to offer a sufficiently discriminative approach and as a result significant research has been dedicated toward identifying bio-markers to guide post-operative surveillance in patients having undergone potential curative surgery. While prognostic models that combine the use of conventional histological parameters and circulating or tumour based biomarkers have been explored in RCC [27], to date none have been prospectively tested for routine clinical use.
In this current work we show both p4E-BP1 and eIF4E to be independent influential prognosticators when analysed as individual biomarkers. More meaningfully we show a significant association between the presence of p4E-BP1 and the over-expression of eIF4E in clinically confined renal tumours of clear cell histology. The direct nature of this association may be corroborated by similar findings in other tumour types [10,28] and by contrast to the lack of correlation between eIF4E and another downstream molecule, pS6, from the mTOR pathway [24]. Specifically, using the same archived patient cohort we have previously reported [24] a lack of correlation between pS6 and the over-expression of eIF4E (P = 0.342). We also found in this present work neither p4E-BP1 or eIF4E alone, or their combined expression, correlated with any specific clinico-pathological parameter for RCC. However, multi-variate analysis demonstrated the combined molecular signature of p4E-BP1/eIF4E to be at least as powerful as either high tumour grade or high tumour stage for predicting the onset of metastatic disease. Collectively, these findings provide compelling evidence that both p4E-BP1 and eIF4E are important functionally-linked determinants of early relapse in ccRCC that are able to identify aggressive disease in a subset of patients, as determined by their significantly shorter DFS. Further, the combined expression of p4E-BP1/eIF4E was able to identify a sub-population of patients that displayed low grade/low stage disease but were associated with a high risk of relapse.
We have previously shown the expression of p4E-BP1 to be significantly associated with clinically confined papillary renal cell carcinoma, a relatively indolent renal tumour subtype where the reported incidence of relapse is much less than for ccRCC [24]. Reasons for the increased prognostic power of p4E-BP1 in renal tumours of clear cell histology most probably reflect other molecular alterations that operate in a concerted manner to drive disease progression. Such candidates may include the increased over-expression of eIF4E, the rate limiting step in protein translation, and the loss of functional VHL. Indeed, it has recently been shown in RCC cell lines that VHL can directly bind pre-40S ribosomal subunits to inhibit ribosome biogenesis and repress global protein synthesis [29]. Therefore, the presence of p4E-BP1, coupled with synergistic over-expression of eIF4E on a VHL mutated genetic background would provide all the necessary machinery for enhanced global protein translation to rapidly drive the progression in ccRCC as reported in this current study.
It has been suggested that cancer cells in general which over-express eIF4E are more dependent upon eIF4E than cancer cells driven by other oncogenes and that this dependence represents an “Achilles heel” [30]. In other tumour types the overexpression of eIF4E has been shown to be dependent upon the presence and/or phosphorylation status of p4E-BP1 [10,28,31], the exact molecular mechanism responsible for the over-expression of eIF4E in RCC is currently unknown. One likely pathway is an interaction between eIF4E and c-myc, the latter a molecule known to overexpressed in advanced RCC [32,33]. Therefore, increased phosphorylation of 4E-BP1 in renal cancer cells will result in the release of eIF4E from its inhibitory conformation leading to the production of oncogenic proteins, which include amongst others c-myc. It is known that the eIF4E gene promoter contains two canonical c-myc response elements that are responsive to transcriptional activation by c-myc in a number of different screens [34]. Any newly formed c-myc therefore has the ability to further potentiate the activation of the eIF4E gene in a positive regulatory feedback loop leading to increasing levels of eIF4E driving disease progression.
Although molecular targeted therapies such as the tyrosine kinase inhibitors (TKIs) e.g. sunitinib or pazopanib, mTOR inhibitors e.g. temsirolimus, or VEGF antibodies e.g. bevacizumab, have significantly improved survival rates for mRCC, not all patients respond favourably and of those that do, drug resistance eventually develops. Molecular pathology is positioned for the discovery and development of tumour biomarkers that are able to predict and monitor treatment response in patients. Indeed the use of combined biomarkers is shown to have greater value than individual biomarkers alone for the prediction of treatment response as well as prognostication in a number of different cancer types, particularly where the biomarkers reflect an intrinsic biologically relevant mechanism or pathway [1]. The p4E-BP1/eIF4E axis we describe in this current study represents a critical convergence point for several upstream signalling pathways such as the EGF-R/ERK and AKT/mTOR, all of which are targeted in some respect by molecular-targeted therapies used in the treatment of renal tumours. Phosphorylated 4E-BP1 has recently been shown to be the single most accurate bio-marker of the mTOR pathway for predicting treatment response in patients with mRCC treated with mTOR inhibitors [35]. However, the above study did not include a sub-analysis of patient eIF4E status which, we propose from our present work would further improve patient discrimination toward treatment stratification options for molecular targeted therapies. This conjecture is further supported by recent pre-clinical studies in renal cancer cells isolated from in vivo tumour models showing that the 4E-BP1/eIF4E axis to be the principal driver of VEGF-A production [36].
In summary, we show that both p4E-BP1 and eIF4E overexpression demonstrates synergy in driving the patho-biology of renal tumours of clear cell origin and when assessed together can accurately predict early relapse in patients with clinically confined disease. Specifically, we show that the combined expression of p4E-BP1 and eIF4E is a biologically relevant biomarker combination that has equal or superior prognostic value than current conventional histological parameters. We propose that the assessed combined expression of p4E-BP1 and eIF4E will have potential to guide and improve patient management with respect to post-operative surveillance and the instigation of targeted therapies. Given that p4E-BP1 and eIF4E are important linked determinants of early relapse in ccRCC, the targeting of the 4E-BP1/eIF4E axis in combination with either cytokine or targeted therapies to prevent or delay relapse appears a rational therapeutic approach [37]. Further examination of the p4E-BP1/eIF4E signature in other patients cohorts is warranted, particularly those having received molecular targeted therapies.
Disclosure of conflict of interest
None.
References
- 1.Stewart GD, O’Mahony FC, Powles T, Riddick AC, Harrison DJ, Faratian D. What can molecular pathology contribute to the management of renal cell carcinoma? Nat Rev Urol. 2011;8:255–265. doi: 10.1038/nrurol.2011.43. [DOI] [PubMed] [Google Scholar]
- 2.Macleod LC, Hotaling JM, Wright JL, Davenport MT, Gore JL, Harper J, White E. Risk factors for renal cell carcinoma in the VITAL study. J Urol. 2013;190:1657–1661. doi: 10.1016/j.juro.2013.04.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smaldone MC, Uzzo RG. Active surveillance: a potential strategy for select patients with small renal masses. Future Oncol. 2011;7:1133–1147. doi: 10.2217/fon.11.97. [DOI] [PubMed] [Google Scholar]
- 4.Goyal R, Gersbach E, Yang XJ, Rohan SM. Differential diagnosis of renal tumors with clear cytoplasm: clinical relevance of renal tumor subclassification in the era of targeted therapies and personalized medicine. Arch Pathol Lab Med. 2013;137:467–480. doi: 10.5858/arpa.2012-0085-RA. [DOI] [PubMed] [Google Scholar]
- 5.Rink M, Chun FK, Robinson B, Sun M, Karakiewicz PI, Bensalah K, Fisch M, Scherr DS, Lee RK, Margulis V, Shariat SF. Tissue-based molecular markers for renal cell carcinoma. Minerva Urol Nefrol. 2011;63:293–308. [PubMed] [Google Scholar]
- 6.Maroto P, Rini B. Molecular biomarkers in advanced renal cell carcinoma. Clin Cancer Res. 2014;20:2060–2071. doi: 10.1158/1078-0432.CCR-13-1351. [DOI] [PubMed] [Google Scholar]
- 7.Hayes DF, Allen J, Compton C, Gustavsen G, Leonard DGB, McCormack R, Newcomer L, Pothier K, Ransohoff D, Schilsky RL, Sigal E, Taube SE, Tunis SR. Breaking a vicious cycle. Sci Transl Med. 2013;5:196cm6. doi: 10.1126/scitranslmed.3005950. [DOI] [PubMed] [Google Scholar]
- 8.Noske A, Lindenberg JL, Darb-Esfahani S, Weichert W, Buckendahl AC, Sehouli J, Dietel M, Denkert C. Activation of mTOR in a subgroup of ovarian carcinomas: correlation with p-eIF-4E and prognosis. Oncol Rep. 2008;20:1409–1417. [PubMed] [Google Scholar]
- 9.Korkolopoulou P, Levidou G, El-Habr EA, Piperi C, Adamopoulos C, Samaras V, Boviatsis E, Thymara I, Trigka EA, Sakellariou S, Kavantzas N, Patsouris E, Saetta AA. Phosphorylated 4Ebinding protein 1 (p-4E-BP1): a novel prognostic marker in human astrocytomas. Histopathology. 2012;61:293–305. doi: 10.1111/j.1365-2559.2012.04236.x. [DOI] [PubMed] [Google Scholar]
- 10.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, Parsons SH, Brail LH, Colligan BM, Koop JW, Hurst BM, Deddens JA, Neubauer BA, Stancato LF, Carter HW, Douglass LA, Carter JH. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69:3866–3873. doi: 10.1158/0008-5472.CAN-08-3472. [DOI] [PubMed] [Google Scholar]
- 11.Jia Y, Polunovsky V, Bitterman PB, Wagner CR. Cap-dependent translation initiation factor eIF4E: an emerging anticancer drug target. Med Res Rev. 2012;32:786–814. doi: 10.1002/med.21260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villalonga P, Fernández de Mattos S, Ridley AJ. RhoE inhibits 4E-BP1 phosphorylation and eIF4E function impairing cap-dependent translation. J Biol Chem. 2009;284:35287–35296. doi: 10.1074/jbc.M109.050120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T, Pelletier J. Eukaryotic initiation factor 4F: a vulnerability of tumor cells. Future Med Chem. 2012;4:19–31. doi: 10.4155/fmc.11.150. [DOI] [PubMed] [Google Scholar]
- 14.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–486. doi: 10.1038/nm1042. [DOI] [PubMed] [Google Scholar]
- 15.Zhou FF, Yan M, Guo GF, Wang F, Qiu HJ, Zheng FM, Zhang Y, Liu Q, Zhu XF, Xia LP. Knockdown of eIF4E suppresses cell growth and migration, enhances chemosensitivity and correlates with increase in Bax/Bcl-2 ratio in triple-negative breast cancer cells. Med Oncol. 2011;28:1302–1307. doi: 10.1007/s12032-010-9630-0. [DOI] [PubMed] [Google Scholar]
- 16.Choi CH, Lee JS, Kim SR, Lee YY, Kim CJ, Lee JW, Kim TJ, Lee JH, Kim BG, Bae DS. Direct inhibition of eIF4E reduced cell growth in endometrial adenocarcinoma. J Cancer Res Clin Oncol. 2011;137:463–469. doi: 10.1007/s00432-010-0902-z. [DOI] [PubMed] [Google Scholar]
- 17.Salehi Z, Mashayekhi F. Expression of the eukaryotic translation initiation factor 4E (eIF4E) and 4E-BP1 in esophageal cancer. Clin Biochem. 2006;39:404–409. doi: 10.1016/j.clinbiochem.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Heikkinen T, Korpela T, Fagerholm R, Khan S, Aittomaki K, Heikkila P, Blomqvist C, Carpen O, Nevanlinna H. Eukaryotic translation initiation factor 4E (eIF4E) expression is associated with breast cancer tumor phenotype and predicts survival after anthracycline chemotherapy treatment. Breast Cancer Res Treat. 2013;141:79–88. doi: 10.1007/s10549-013-2671-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang XL, Cai HP, Ge JH, Su XF. Detection of eukaryotic translation initiation factor 4E and its clinical significance in hepatocellular carcinoma. World J Gastroenterol. 2012;18:2540–2544. doi: 10.3748/wjg.v18.i20.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Fan S, Koo J, Yue P, Chen ZG, Owonikoko TK, Ramalingam SS, Khuri FR, Sun SY. Elevated expression of eukaryotic translation initiation factor 4E is associated with proliferation, invasion and acquired resistance to erlotinib in lung cancer. Cancer Biol Ther. 2012;13:272–280. doi: 10.4161/cbt.18923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martineau Y, Azar R, Bousquet C, Pyronnet S. Anti-oncogenic potential of the eIF4E-binding proteins. Oncogene. 2013;32:671–677. doi: 10.1038/onc.2012.116. [DOI] [PubMed] [Google Scholar]
- 22.Griffiths DFR, Verghese A, Golash A, Kynaston HG, Matthews PN, Hart AJL, Court JB. Contribution of grade, vascular invasion and age to outcome in clinically localized renal cell carcinoma. BJU Int. 2002;90:26–31. doi: 10.1046/j.1464-410x.2002.02661.x. [DOI] [PubMed] [Google Scholar]
- 23.Campbell L, Gumbleton M, Griffiths DFR. Caveolin-1 overexpression predicts poor disease-free survival of patients with clinically confined renal cell carcinoma. Br J Cancer. 2003;89:1909–1913. doi: 10.1038/sj.bjc.6601359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell L, Jasani B, Edwards K, Gumbleton M, Griffiths DFR. Combined expression of caveolin-1 and an activated AKT/mTOR pathway predicts reduced disease-free survival in clinically confined renal cell carcinoma. Br J Cancer. 2008;98:931–940. doi: 10.1038/sj.bjc.6604243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuhrman SA, Lasky LC, Limas C. Prognostic significance of morphologic parameters in renal cell carcinoma. Am J Surg Pathol. 1982;6:655–663. doi: 10.1097/00000478-198210000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Thomas DH, Verghese A, Kynaston HG, Griffiths DF. Analysis of the prognostic implications of different tumour margin types in renal cell carcinoma. Histopathology. 2003;43:374–380. doi: 10.1046/j.1365-2559.2003.01721.x. [DOI] [PubMed] [Google Scholar]
- 27.Finley DS, Pantuck AJ, Belldegrun AS. Tumor biology and prognostic factors in renal cell carcinoma. Oncologist. 2011;16(Suppl 2):4–13. doi: 10.1634/theoncologist.2011-S2-04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coleman LJ, Peter MB, Teall TJ, Brannan RA, Hanby AM, Honarpisheh H, Shaaban AM, Smith L, Speirs V, Verghese ET, McElwaine JN, Hughes TA. Combined analysis of eIF4E and 4E-binding protein expression predicts breast cancer survival and estimates eIF4E activity. Br J Cancer. 2009;100:1393–1399. doi: 10.1038/sj.bjc.6605044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao WT, Zhou CF, Li XB, Zhang YF, Fan L, Pelletier J, Fang J. The von Hippel-Lindau protein pVHL inhibits ribosome biogenesis and protein synthesis. J Biol Chem. 2013;288:16588–16597. doi: 10.1074/jbc.M113.455121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borden KL. Targeting the oncogene eIF4E in cancer: From the bench to clinical trials. Clin Invest Med. 2011;34:E315. doi: 10.25011/cim.v34i6.15889. [DOI] [PubMed] [Google Scholar]
- 31.Seki N, Takasu T, Sawada S, Nakata M, Nishimura R, Segawa Y, Shibakuki R, Hanafusa T, Eguchi K. Prognostic significance of expression of eukaryotic initiation factor 4E and 4E binding protein 1 in patients with pathological stage I invasive lung adenocarcinoma. Lung Cancer. 2010;70:329–334. doi: 10.1016/j.lungcan.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi S, Yoshihiro S, Matsuyama H, Nagao K, Fukunaga K, Matsumoto H, Matsuda K, Oba K, Naito K. The allelic loss of chromosome 3p25 with c-myc gain is related to the development of clear-cell renal cell carcinoma. Clinical Genetics. 2003;63:184–191. doi: 10.1034/j.1399-0004.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 33.Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, Hsu CI, Lin WC, Lai MK, Lin JY. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. 2009;273:35–43. doi: 10.1016/j.canlet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt EV. The role of c-myc in regulation of translation initiation. Oncogene. 2004;23:3217–3221. doi: 10.1038/sj.onc.1207548. [DOI] [PubMed] [Google Scholar]
- 35.Nishikawa M, Miyake H, Harada KI, Fujisawa M. Expression level of phosphorylated-4E-binding protein 1 in radical nephrectomy specimens as a prognostic predictor in patients with metastatic renal cell carcinoma treated with mammalian target of rapamycin inhibitors. Med Oncol. 2014;31:792. doi: 10.1007/s12032-013-0792-4. [DOI] [PubMed] [Google Scholar]
- 36.Dodd KM, Yang J, Shen MH, Sampson JR, Tee AR. mTORC1 drives HIF-1α and VEGF-A signalling via multiple mechanisms involving 4EBP1, S6K1 and STAT3. Oncogene. 2014;34:2239–2250. doi: 10.1038/onc.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Attar-Schneider O, Drucker L, Zismanov V, Tartakover-Matalon S, Rashid G, Lishner M. Bevacizumab attenuates major signaling cascades and eIF4E translation initiation factor in multiple myeloma cells. Lab Invest. 2012;92:178–190. doi: 10.1038/labinvest.2011.162. [DOI] [PubMed] [Google Scholar]