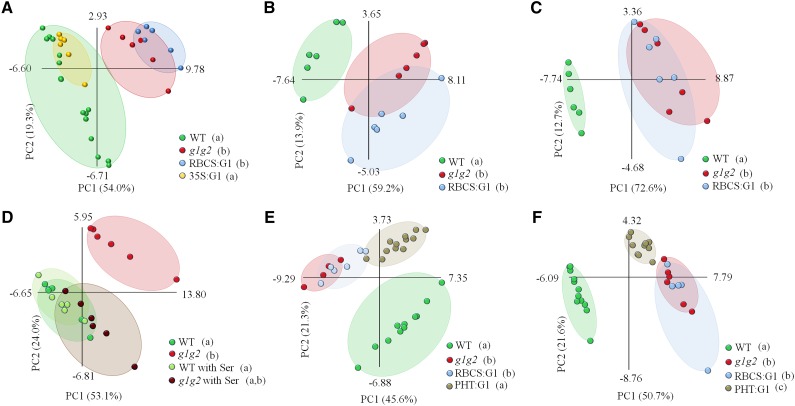
Figure 2.
PCA of the metabolite profiles of different GAPCp mutant lines. Data from the gas chromatography-mass spectrometry analysis were evaluated using PCA, with the two first components accounting for at least 67% of total metabolic variance. Values in parentheses give the relative contribution of each component to the total variance observed in the data set. The first principal component was also used for the ANOVA of different lines, with Tukey’s posthoc tests to differentiate between the lines. A, The AP at the end of the light period. According to Tukey’s test, two different subsets are established: subset 1 (wild type [WT] and gapcp1gapcp2 35S:GAPCp1 [35S:G1] [a]) and subset 2 (gapcp1gapcp2 [g1g2] and g1g2 RBCS:GAPCp1 [RBCS:G1] [b]). B, The AP at the end of the dark period. According to Tukey’s test, two different subsets are established: subset 1 (wild type [a]) and subset 2 (g1g2 and RBCS:G1 [b]). C, The AP at the end of the light period in medium supplemented with 2% (w/v) Suc. According to Tukey’s test, two different subsets are established: subset 1 (wild type [a]) and subset 2 (g1g2 and RBCS:G1 [b]). D, The AP at the end of the light period in medium supplemented with 0.1 mm Ser. According to Tukey’s test, two different subsets are established: subset 1 (g1g2 with Ser, wild type, and wild type with Ser [a]) and subset 2 (g1g2 and g1g2 with Ser [b]). E, The AP at the middle of the light period. According to Tukey’s test, two different subsets are established: subset 1 (wild type and g1g2 PHT:GAPCp1 [PHT:G1] [a]) and subset 2 (g1g2 and RBCS:G1 [b]). F, Roots at the middle of the light period. According to Tukey’s test, three different subsets are established: subset 1 (wild type [a]), subset 2 (g1g2 and RBCS:G1 [b]), and subset 3 (PHT:G1 [c]). Each ellipse includes all the data points of the same line being analyzed (cover hull).