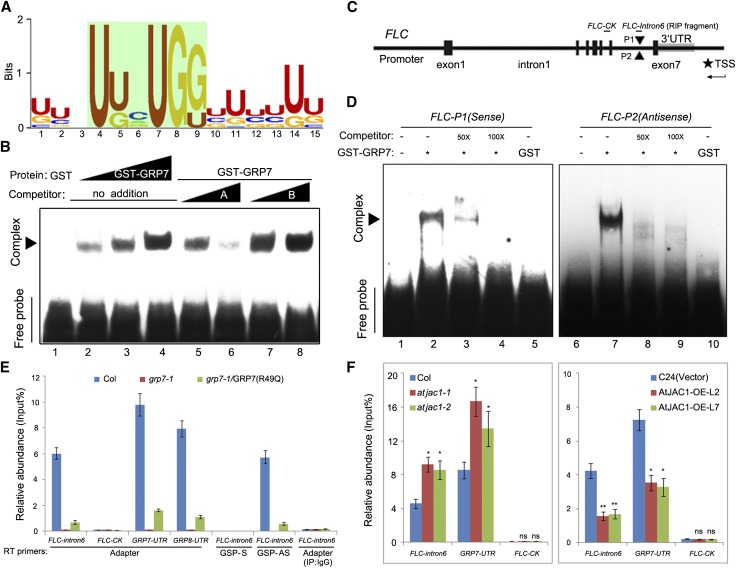
Figure 6.
GRP7 directly binds FLC antisense premRNA through a conserved motif. A, The most significantly enriched RNA-binding motif of GRP7 screened by SELEX and analyzed by MEME2. B, RNA-EMSA to confirm the binding of GRP7 to the motif screened out by SELEX in vitro. Competitor A was a non-biotin-labeled GRP7-binding motif, and competitor B was a nonrelevant RNA fragment without biotin labeling. Sequences of individual oligonucleotides are listed in the supplemental sheet. C, Schematic diagram of the positions of putative GRP7-binding motifs in the FLC genomic sequence. Gene structure is as indicated. P1 and P2, Two putative GRP7-binding sites in FLC intron 6 with opposite directions; TSS, transcription start site for antisense FLC. D, GRP7 binds to FLC sense and antisense RNA probes in RNA-EMSA in vitro. Competitors are non-biotin-labeled RNA fragment FLC-P1 or FLC-P2. E, RIP followed by qPCR to confirm the binding of GRP7 to antisense FLC premRNA in vivo. FLC-intron6, FLC sense or antisense premRNA fragment containing putative GRP7-binding sites, with position indicated in C; FLC-control check fragment (FLC-CK), FLC premRNA fragment containing no GRP7-binding sites as a negative control, with position indicated in C; GSP, gene-specific primer. The GRP7/GRP8-UTR fragment served as a positive control, and grp7-1 transformed with pGRP7:GRP7(R49Q) served as a negative control for GRP7-binding activity. F, The binding of GRP7 to FLC antisense premRNA was altered in atjac1 mutants and AtJAC1-OE plants compared with controls. Adaptor primers were used for RT after determining the RNA pool with IP. Data shown in E and F are means ± sd of three parallel samples from one replicate, and three biological repeats were performed. Statistical analysis was done by Student’s t test: *, P < 0.05; **, P < 0.01; ns, no significant change.