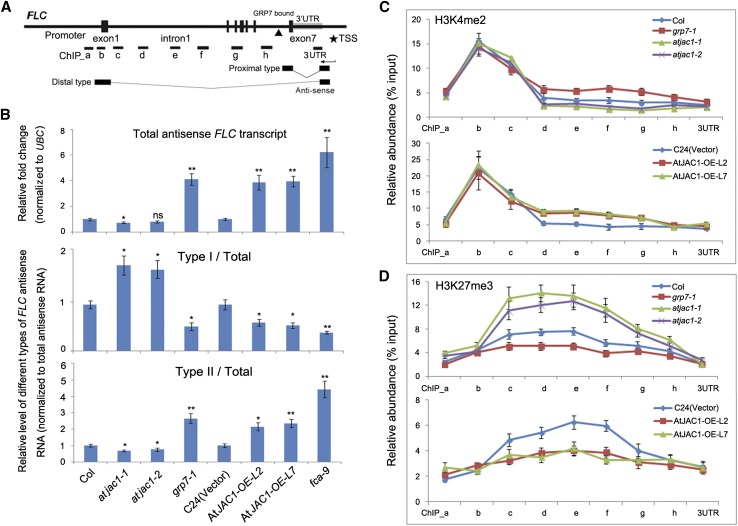
Figure 7.
GRP7 and AtJAC1 influence the RNA processing of COOLAIR and histone modification within the FLC gene body. A, Diagram of FLC gene structure and antisense transcripts. The black triangle indicates the GRP7-binding site. PCR-amplified fragments for chromatin immunoprecipitation (ChIP) in C and D are indicated as ChIP_a to 3UTR. B, Relative expression of total antisense mRNA (normalized to UBC) as well as relative ratio of proximal (type I) and distal (type II) antisense transcripts (normalized to total antisense transcripts) in atjac1, grp7-1, and fca-9 mutants and AtJAC1-OE plants compared with controls. In total, three biological repeats were performed. Statistical analysis was done by Student’s t test; atjac1 mutants, grp7-1, and fca-9 were compared with Col, whereas AtJAC1-OE lines were compared with C24 (vector): *, P < 0.05; **, P < 0.01; ns, no significant change. C and D, Histone modification change (H3K4me2 [C] and H3K27me3 [D]) at the FLC locus in atjac1, grp7-1 mutants, and AtJAC1-OE plants compared with controls. The y axis shows the relative abundance of ChIP sample compared with input. Data are means ± sd of three parallel samples from one replicate, and three biological repeats were performed.