Figure 2.
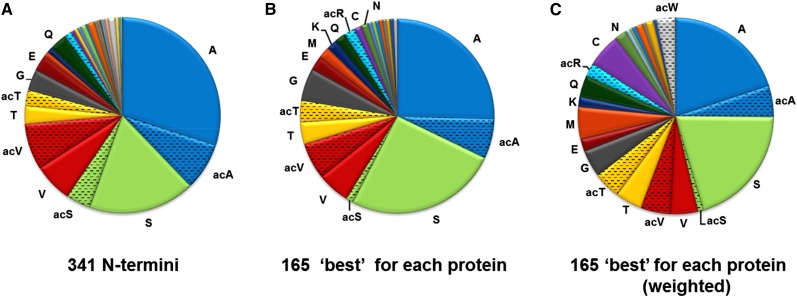
Nt amino acid frequency for stroma-exposed n-encoded chloroplast proteins. A, All detected stromal Nti (341), excluding unprocessed proteins and obvious breakdown products (Supplemental Table S4). This shows that Ala and Ser are heavily favored as Nt residues, followed by Val and Thr, while 14 residues were underrepresented (Gly, 14×; Gln, 14×; Glu, 10×; Ile, 6×; Arg, 5×; Lys, 5×; Met, 4×; Asn, 3×; Leu, 3×; Cys, 2×; Phe, 2×; Trp, 1×; Tyr, 1×; and Asp, 1×) or not observed (Pro and His). A significant portion of these highly favored residues were acetylated (Val, 54%; Thr, 47%; Ala, 21%; and Ser, 19%), whereas the acetylation rate for other residues was either 0% (Tyr, Leu, Phe, Asp, and Cys) or 100% (Trp; acetylation is indicated as ac). B, Single highest ranked N terminus per protein (165), excluding Nti with less than two SPC. Selecting a single best or highest ranked N terminus for each protein (see “Materials and Methods”) hardly influenced the Nt amino acid frequency, except that it slightly decreased the dominance of Ala, increased Ser, and reduced acetylated Ser. Less frequent residues were Gly (9×), Glu (5×), Gln (4×), Lys (4×), Arg (3×), Met (3×), Asn (2×), Cys (2×), Leu (1×), Ile (1×), Trp (1×), Tyr (1×), and Asp (1×), whereas Phe, Pro, and His were not observed. C, Single highest ranked Nti as in B but normalized (weighted) to the frequency of each amino acid in the known (from the Plant Proteome Data Base [PPDB]; 1,575 proteins) n-encoded plastid proteome with predicted cTPs removed.