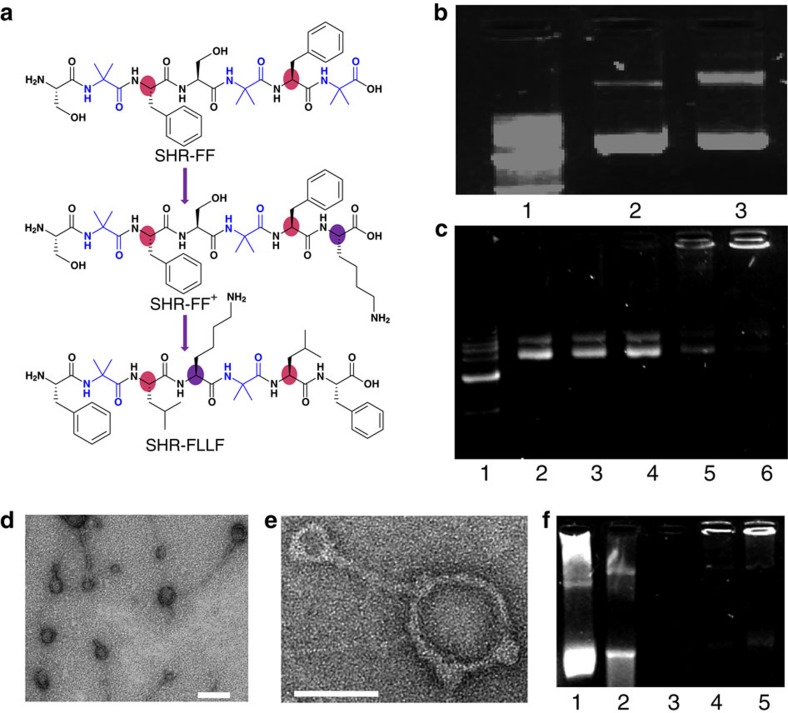
Figure 4. Structural modification and DNA condensation by modified single heptad peptide.
(a) Modification of the SHR-FF sequence to afford the positively charged single heptad repeat module containing single lysine residue. Colour regions display the significant variation in amino-acid residues. (b) Agarose-gel electrophoretic assay of DNA condensation by SHR-FF+. Lane 1: 1 kb marker; lane 2: free DNA; lane 3: N/P 100. (c) Agarose-gel electrophoretic assay of DNA condensation by SHR-FLLF. Lane 1: 1 kb marker; lane 2: free DNA; lane 3: N/P 1; lane 4: N/P 20; lane 5: N/P 50: lane 6; N/P 100. (d) TEM images of complexes of plasmid DNA with SHR-FLLF (N/P 100). Scale bar, 100 nm. (e) Magnified TEM image showed the presence of rod-like and toroidal structures of a peptide-DNA complex. Scale bar, 50 nm. (f) DNase I digestion assay. Lane 1: free DNA without DNase I; lanes 2–3: free DNA treated with DNase I for 5 and 30 min, respectively; lanes 4–5: DNA-peptide (N/P 100) complex treated with DNase I for 5 and 30 min, respectively (N/P is the ratios between positive charges of peptide to negative charges of phosphate groups in DNA).