Abstract
The panzootic of H5N1 influenza in birds has raised concerns that the virus will mutate to spread more readily in people, leading to a human pandemic. Mathematical models have been used to interpret past pandemics and outbreaks, and to thus model possible future pandemic scenarios and interventions. We review historical influenza outbreak and transmission data, and discuss the way in which modellers have used such sources to inform model structure and assumptions. We suggest that urban attack rates in the 1918–1919 pandemic were constrained by prior immunity, that R 0 for influenza is higher than often assumed, and that control of any future pandemic could be difficult in the absence of significant prior immunity. In future, modelling assumptions, parameter estimates and conclusions should be tested against as many relevant data sets as possible. To this end, we encourage researchers to access FluWeb, an on‐line influenza database of historical pandemics and outbreaks.
Keywords: Communicable disease control, disease outbreaks, disease transmission, influenza A virus, H5N1 subtype, historical cohort studies, mathematical model
Introduction
Recent outbreaks of H5N1 influenza among domesticated poultry in South‐East Asia have had unprecedented economic impacts. 1 , 2 Seasonal migration of infected wild birds and trade in live poultry 3 have spread H5N1 viruses throughout Asia, Europe and Africa. Direct transmission of this avian virus to humans through contact with infected live birds, poultry products or excreta had caused 154 confirmed human deaths by November 2006, 4 with updates posted on the World Health Organization (WHO) website since that time (http://www.who.int/csr/disease/avian_influenza/updates/en/index.html). Earlier reports of suspected secondary infection in family members 1 , 5 have been supported by recent evidence of human‐to‐human transmission of an H5N1 mutant within a household. 6 In the absence of prior immunity, the emergence of novel virus that is more readily transmissible could cause a human pandemic to rival or surpass the pandemics of 1889–1891, 7 1918–1919 (H1N1), 1957 (H2N2) and 1968 (H3N2). 1 Public health bodies and governments have sought urgent advice in planning how to respond to this new threat. 8
Why bother with mathematical models?
The pandemic of 1918–1919 was characterized by high attack rates, several waves of infection and high mortality in young adults. Such observations can be better understood by fitting explanatory models to the data to estimate R 0 and other parameters governing influenza spread. 9 , 10 These parameter estimates can then be used to simulate model scenarios to examine the possible consequences of proposed outbreak control strategies. 11
In an outbreak in a fully susceptible population, the average number of secondary cases infected by a primary case is denoted by the reproduction number, R 0, which depends on the duration of the infectious period (1/γ), and a transmission parameter (β), influenced by viral and host factors and contact opportunities. In its simplest form:
R 0 can be estimated from the final attack rate if the population was fully susceptible at the outset. However, in many situations, populations have pre‐existing immunity, and a proportion of exposures leads to subclinical infections, reducing the apparent attack rate. The proportion immune (ω) within a particular population determines the effective reproductive number (R) by the relationship:
R 0 provides a worst‐case scenario for the attack rate. Furthermore, in combination with the mean generation time (serial interval between cases) and the proportion susceptible, R 0 determines the rate of spread of an outbreak. In favourable circumstances, R 0 can be estimated from outbreak data, with parallel inferences about levels of subclinical infection and prior immunity.
This article reviews aspects of influenza biology and epidemiological findings from past influenza epidemics. It provides insight into the nature of influenza infection in individuals, and the dissemination of virus in households, schools and populations. We highlight the importance of susceptibility, pre‐existing immunity and subclinical infection in modifying the apparent attack rate and spread of influenza, and suggest that mathematical models of influenza transmission would be improved if they could account more adequately for such factors. Finally, we review the potential utility of interventions such as social distancing, antivirals and immunization to prevent or limit outbreaks.
Biology and epidemiology of influenza infections
Influenza viruses probably evolved in wild waterfowl, and then spread to other animals, including humans. 12 In order to successfully cross the species barrier, mutant strains had to acquire the capability to bind the dominant receptor in the human respiratory epithelium, which differs from that in birds. 12 , 13 , 14 Mutation of the virus sufficient to allow such binding could produce an H5N1 strain able to spread more effectively from person to person, potentially triggering a new pandemic. 15
Inter‐pandemic influenza affects 5–30% of the population each winter in temperate climes. Attack rates fall with age, presumably because of incremental immunity following repeated exposures, but rise again in the elderly. Mortality rates are generally low, but higher at extremes of age. As inter‐pandemic viruses circulate in partially immune populations, haemagluttinin (HA) antigens mutate quickly to escape antibody directed against earlier viruses, a phenomenon known as ‘antigenic drift’. 12
T‐cell immunity is also important; cytotoxic T‐cells kill virus‐infected epithelial cells to clear existing infections. If protective antibody is minimal, as with a new pandemic strain, many epithelial cells may become infected, and a hyperactive cytotoxic response against these cells can cause lung destruction and death. This scenario could explain the higher mortality for young adults during the 1918–1919 pandemic. 12 , 16
Immunity and susceptibility to infection
Some hosts are more susceptible to influenza because they have little or no acquired immunity or because of intrinsic factors affecting innate immunity. 12 , 17 Isolated populations with no recent influenza exposure are particularly vulnerable, as with Native Americans and Pacific Islanders during the 1918–1919 pandemic. 18 , 19 Likewise, on Tristan da Cunha, where influenza had been absent for many years, the first arrival of H3N2 virus by ship from Cape Town in 1970 led to 96% of persons falling ill, with repeat attacks in a significant minority. 20
Consistent with the premise of short‐lived protection through past exposures to inter‐pandemic influenza, much lower attack rates were reported in urbanized populations during the 1918–1919, 1957 and 1968 influenza pandemics. In Cleveland in 1957, 47% of household members had serologically confirmed influenza during a 10‐week period. Disease incidence in children aged 10–14 years was three times that in adults, in keeping with their relative naivety to influenza. 21 , 22 In contrast, more uniform attack rates were observed up to 50 years of age in the 1968 H3N2 pandemic, with relative sparing of people born before 1918 suggesting that exposure to related viral antigens more than 50 years earlier had conferred lasting protection. 23 , 24 , 25 , 26
In the Houston family study of 1976, high titres of subtype‐specific haemagglutination‐inhibiting (HI) antibody titres protected against H3N2 influenza. Breakthrough infections did occur in those with high titres, but were usually milder. 27 In the Seattle cohort study from 1975 to 1979, HI antibody thresholds conferring protection differed for the three strains circulating over the period: A/H3N2, A/H1N1 and B. 28 , 29 Such variation is likely due to differences in the immunogenicity of HA antigens. 30 Further, attack rates in Seattle children were double those seen in adults for a given HI titre, 29 suggesting the importance of unmeasured humoral and cellular immunity. 22 , 31 , 32 Broadly reactive ‘recall’ immune responses, seen within days of vaccination, 33 could explain the rarity of H1N1 infection in Seattle adults with low HI titres and a cohort history of exposure to a related strain 26 years earlier. 28
Models incorporating immunity
Spicer and Lawrence modelled influenza in Greater London from 30 years of mortality data. Exposure to successive drift mutants was assumed to reduce the susceptible pool, which was replenished by births and the emergence of antigenically novel strains. 34 In a model for South‐East Asia, Ferguson et al. assumed that 27% of rural households would be resistant to a novel strain of influenza as a result of recent contact with related antigens. 10 Other models allow the risk of transmission, 11 , 35 or progression to disease given acquisition, 36 , 37 to depend on age, indirectly incorporating immune protection. Longini et al. explicitly calculated the influence of ‘high’ or ‘low’ antibody titres on influenza risk in children and adults from epidemic data, but could not refine their estimates. 38
The simplest influenza models assume that a single episode of infection confers lifelong immunity. This cannot be true, especially for children, who can experience three or four clinical episodes in successive seasons. 28 Ferguson et al. modelled the combined effects of waning immunity and antigenic drift for inter‐pandemic virus by assuming that a single exposure to influenza protected against related strains for 5 years, after which individuals were again fully susceptible. Less robust immune responses in the very young and elderly were modelled by shortening this duration of protection. 37
In the absence of priming, immune responses to novel antigens can be insufficient to confer short‐term protection, even in adults. This could explain the second wave of infections observed on Tristan da Cunha, in which 33% of islanders experienced a second (usually milder) attack within a month of the initial outbreak. 20 Repeat infections after a short interval have been reported in other outbreaks, as in the naval apprentice school at Greenwich in 1924 (Figure 1) 39 and the summer, autumn and winter waves of the 1918–1919 pandemic in England. 19
Figure 1.
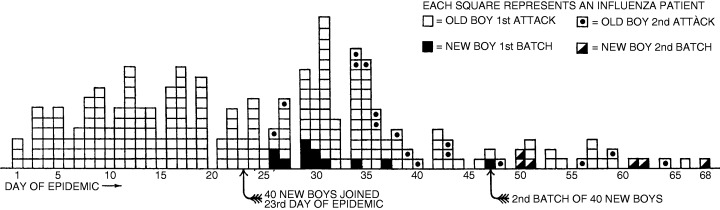
The 1924 influenza epidemic at the Royal Naval School, Greenwich. The introduction of new classes of (susceptible) students led to recrudescence of the outbreak with both ‘new boys’ and ‘old boys’ affected. 39
Multiple‐wave data from Tristan da Cunha and from the 1918–1919 pandemic in Royal Air Force camps in the UK (Figure 2) have been modelled to estimate R 0 as well as parameters for prior immunity, waning of immunity/antigenic drift, and subclinical infection, with consistent results (J. Mathews, personal communication).
Figure 2.
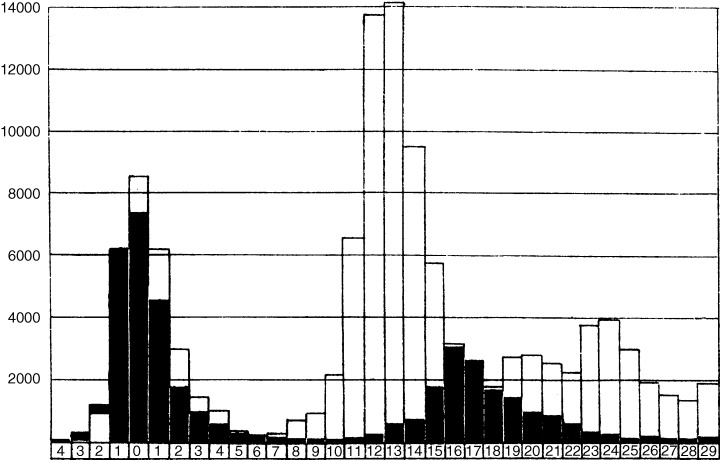
Incidence of influenza cases in Royal Air Force camps (black columns) and the city of Copenhagen (white columns) during the 1918–1919 pandemic. 19 Time‐line units are in weeks measured from the maximum of the initial peak. Both curves display multiple waves, consistent with hypotheses of waning immunity and/or antigenic drift. De‐mobilization at the conclusion of World War I could have prevented the third wave, visible for Copenhagen, from being registered in the Air Force camps.
Infectivity, transmission and subclinical infection
After experimental inoculation of adults with influenza, virus titres in nasal lavage fluid rise at 24 hours, peak with fever onset at 48 hours, and then decline over the next 5 days. 40 , 41 In children with natural infections, viral titres continue to rise 24–48 hours after symptom onset. 42 In one community‐based study, 8·3% of children were still shedding influenza virus into the second week of illness. 43 Elderly and immunocompromised patients can shed virus for weeks or even months. 44 Conversely, of cases detected by seroconversion during epidemic influenza seasons, perhaps a third are asymptomatic 29 , 45 and of uncertain infectious potential.
Viral shedding is an imperfect guide to the risk of transmission. During influenza outbreaks, some infected individuals never infect anyone else, whereas in exceptional circumstances as many as 40 secondary cases have been reported. 46 R 0 thus represents a value averaged over different hosts and environments. 47 Experimental studies of influenza acquisition in humans 48 and animals 49 cannot capture subtle effects such as confinement on aeroplanes, 46 seasonal climatic variation 50 or altered conditions during wars. 51
Epidemiological and household studies can provide insights into host factors influencing infectiousness. Pre‐school and school age children are more likely than other infected household members to produce secondary cases among contacts, 52 a phenomenon attributed to density rather than to the duration of shedding in modelling analyses. 53 Accordingly, influenza models incorporating age structure 36 , 54 do not explicitly allow for longer infectious periods in children. Although asymptomatic individuals are unlikely to be highly infectious, serological evidence of ‘off season’ transmission in families has been reported. 29 Models that allow for reduced infectiousness of asymptomatic cases have halved the infectious period 55 or lowered the probability of transmission. 10 , 56 Whether or not ‘severe’ cases shed more virus and are thus more infectious is questionable, as the host immune response plays a significant role in pathogenicity. 16 , 57 , 58
Presymptomatic transmission
Viral shedding studies suggest that transmission could peak soon after the onset of illness in the index case. Ferguson et al. 37 and Fraser et al. 59 estimated that 30–50% of transmission occurred before symptom onset, assuming that illness would result in withdrawal to home. Some models incorporating household structure extend this idea, allowing continued exposure of the index case to family members while reducing community spread. 36 , 56 This assumption has significant consequences for outbreak control, which will be more difficult if most transmission occurs before index cases can be identified. 59
Infectious period, serial interval and latent period
Observational studies of primary and secondary infections in households provide information about serial interval between cases. A mode of 2 days between the onset of symptoms in successively infected household members was reported by medical officers during pandemics in 1890 7 and 1918–1919. 19 The mean serial interval for Kelley's Island households in 1920 was 2·9 days, with a mode of 2 days (Figure 3). 60
Figure 3.
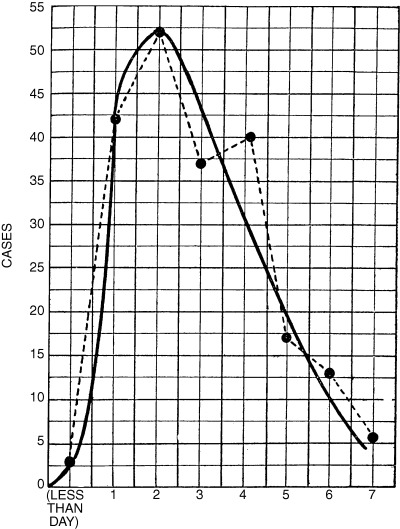
Distribution of the serial interval during the 1920 influenza epidemic on Kelleys Island, Ohio, USA. Interval taken to be the time in days between the first and subsequent cases in a household, assuming that the first case is the source of infection for other household cases. 60
Later household studies during the 1957 pandemic in Cleveland and the 1968 pandemic in Kansas reported median serial intervals of 3 61 and 8 days 23 respectively. However, a mode of 2 days was again seen in Kansas, 23 and in subsequent studies of interpandemic influenza, 52 showing that the median is sensitive to censoring of the observation period. The average serial interval between cases could be longer for transmissions occurring in the community, if there is continuing access to susceptibles.
Elveback et al. 36 and Longini et al. 35 , 56 , 62 separate latent (1·9 days) and infectious (4·1 days) phases of influenza infection, with no transmission during the latent period. 56 Ferguson et al. allow for a similar mean latent phase of 1·48 days in their individual‐based models, but shorten the duration of peak infectiousness to give a serial interval of 2·6 days. 10 , 11 More recent work from the Longini group assumed a slightly longer generation time of 3·5 days. 63
Assumptions about mixing
Opportunities for disease transmission differ by social situation, and location, as seen in the Kelley's island outbreak of 1920 (Figure 4). 60
Figure 4.
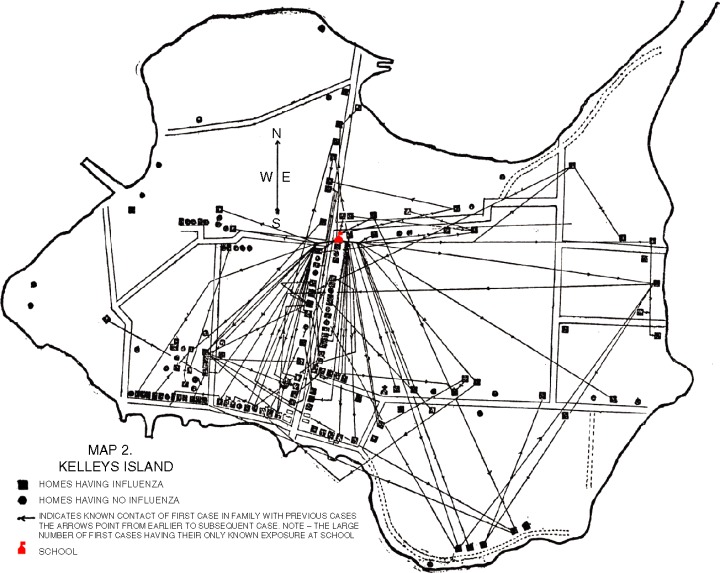
Map showing the location of influenza cases on Kelleys Island, Ohio, USA, during the 1920 epidemic. 60 In this small population, relatively isolated from the outside world, it was possible to reconstruct likely networks and hubs of disease transmission. Paths and arrows indicate a known contact between a first case in a family and a previous case. Note the large number of first cases having their only known exposure at the island's school (indicated).
Hope‐Simpson estimated a fourfold increase in influenza incidence among general practice patients exposed to a household case. 24 Children typically experience higher age‐specific seasonal influenza attack rates than adults, often acquiring the infection from peers in day care centres, kindergartens and day schools. 25 For this reason they more frequently introduce illness into the family, 27 , 29 although in some previous pandemics an adult was as likely to be the household index case as was a child. 23 , 64 Family size and measures of crowding can further influence influenza risk. 27 , 60 , 64 Other residential settings in which large populations of susceptibles may become rapidly infected include boarding schools 19 , 65 and aged care homes. 66 , 67
Indirect evidence about the role of social mixing in disease transmission is provided by the natural history of the 1919 influenza pandemic in Sydney. Following diagnosis of the first cases in late January, government acted to limit spread by closing institutions and meeting places and by requiring masks to be worn in public. Incident cases declined by mid‐February, and restrictions were then removed. A second epidemic wave followed in early March, after which regulations were reimposed. 68 Records from 1919 also reveal higher infection rates in occupations involving travel and frequent contact with the public. 68
Compartmental models often stratify individuals by age to help capture the heterogeneity of contact opportunities in populations. 69 Ferguson et al. employ this method to characterize close mixing in schools and aged care institutions. 37 Longini et al. developed an individual‐based model of a ‘typical’ American population 36 with 1000–2000 people in households of different sizes and age structures, with some attending day care centres and schools. 62 The probability of acquiring infections from the community has been estimated from epidemiological data, 70 with parameters refined to incorporate the influence of age 54 and prior immunity. 38 Analogous models for influenza spread in rural South‐East Asia 10 , 35 and the developed world 11 , 63 have characterized community mixing patterns in a range of social settings. 35 Network type models consider explicit interactions between discrete nodes, with contact probabilities related to social rather than geographical distance. 71 , 72 Other models have simulated spread within and between subpopulations to estimate the probability of local extinction of an outbreak, and subsequent reintroduction. 73 , 74 Incorporation of geographical distance and travel information has allowed modelling of the effects of travel on regional, 75 national 76 and international 76 influenza spread.
Modelling interventions
We discuss modelling approaches to three public health strategies that can limit influenza transmission: social distancing, use of antivirals and immunization.
Social distancing
Social distancing encompasses measures that limit person‐to‐person spread of infection. In the 1918–1919 pandemic, stringent quarantine of the island populations of American Samoa and Australia was of benefit, 18 , 19 although unlikely to be practicable in our age of international air travel. 11 , 77 Restrictions on social movement within communities in 1919 also appeared to limit disease spread. 68 A caveat is that self‐imposed behavioural changes can influence the apparent efficacy of government interventions during any pandemic. 10 , 63
The effects of school closures on community‐wide influenza transmission are not well defined. During the 1920 influenza epidemic on Kelley's island, Ohio, school contacts were found to be important in disease spread. Further, a decline in incidence followed school closure (Figure 5). 60
Figure 5.
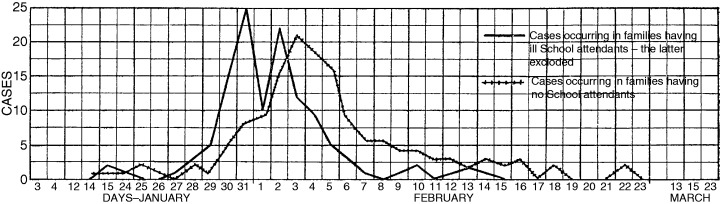
Daily incidence of influenza cases on Kelleys Island, Ohio, USA, during the 1920 epidemic for (1) cases occurring in families having school attendees and (2) cases occurring in families having no schoolchildren. The peak incidence for (1) occurs 3 days before the peak for (2) showing the effect of mixing in school on earlier transmission to households. The island's school was closed on 31 January. 60
More recently, a 30% reduction in paediatric visits to healthcare providers coincided with a 2‐week teachers’ strike during the epidemic influenza season in Israel. 78 Historically, the likely benefit of closing schools was thought to depend on the household living conditions to which children would be returned. 19 Accordingly, models exploring this intervention are sensitive to assumptions about age‐specific transmission rates in the school compared with rates in the home. 11 , 63
Transmission from infected individuals can be reduced by institutional or household quarantine. Household‐based models of this strategy show a fall in effective reproduction rate, contingent upon the timeliness of tracing, the proportion of cases and contacts identified and their compliance. 11 , 79 Infection control measures, including hand hygiene and personal protective equipment, can also reduce the individual risk of respiratory infections. 80 , 81 Although aerosol spread of influenza can occur, 46 its importance, relative to droplet infection, is still controversial, with implications for the standard of protective mask needed to reduce exposure risk. 82 , 83 Importantly, experience of the recent SARS outbreak has shown that well co‐ordinated institutional responses can protect healthcare workers far more than would be predicted from the additive effects of individual measures. 81
Antivirals
The neuraminidase inhibitors (NAIs) zanamivir and oseltamivir antagonize the action of neuraminidase (NA), thereby impairing release of virions from host cells. 84 Although effective in prophylaxis and in reducing the severity and duration of symptoms in interpandemic influenza, the use of these drugs has produced little benefit in H5N1 influenza, 85 possibly because of treatment delay, 85 which reduces efficacy, 86 or higher viral loads. 87
Data from clinical trials with interpandemic influenza show that NAI treatment within 48 hours of symptom onset in virologically confirmed influenza reduces viral shedding by 40–70%. 85 Effects begin within 24 hours, 88 with total suppression of virus achievable by day 2–3 of illness. 89 NAIs have less impact on shedding in children, 90 but a 1‐day reduction in symptom duration is achievable regardless of age. 91 , 92 Prophylaxis with NAIs can reduce the risk of infection in household contacts, measured by seroconversion or viral isolation, over and above treatment of the index case, 93 with efficacy of 60–80%. 85 Breakthrough cases on prophylaxis are less symptomatic. 93
Published models of scenarios using antivirals to limit the spread of pandemic influenza have used estimates of prophylactic efficacy based on such data. 10 , 11 , 55 , 56 , 63 The maximal achievable reduction in infectiousness as a result of treatment has been estimated at 28%. 10 Models allow exploration of a range of targeted scenarios for antiviral use, from treatment of cases and household contacts, 11 to prophylaxis of institutions or geographical regions where disease has been identified. 10 , 35 , 63 The importance of early case detection is paramount, 11 with the size of the antiviral stockpile as another major constraint. In the containment phase of a pandemic, an aggressive combined approach to treatment and prophylaxis of incident cases represents optimal use of a limited resource. 94
Influenza virus resistance to oseltamivir is of potential concern, 95 but at present can be demonstrated in less than 0.5% of strains worldwide. 96 Mutants arising in vitro can have mutations in both HA and NA genes, 97 whereas those from treated immunocompetent patients show single mutations in the NA region. 98 Transmissibility of most resistant strains is greatly reduced, 99 but exceptions occur, 49 leaving no room for complacency. 100 Nine of fifty children treated with oseltamivir carried resistant strains within 4 days, with persistent shedding of both resistant and wild‐type virus to day 7, 97 possibly related to the high viral loads observed in childhood. 101 The rarity of clinical zanamivir resistance 102 may relate to the poor in vitro viability of strains with this mutation. 103
Models exploring the population impact of antiviral resistance confirm that the transmissibility of resistant strains, relative to wild type, and the rate of emergence of such strains are most important as determinants of outcome. 37 Intriguingly, a high rate of production of poorly transmissible mutants could aid, rather than hinder, outbreak control (J. McCaw, personal communication). Models comparing the likely impact of treatment or prophylaxis on the emergence of drug resistance are critically dependent on underlying assumptions. If as much as half of all transmission occurs before case detection, prophylaxis‐based strategies favour selection of mutant strains, 37 whereas if transmission continues for several days after case detection, treatment has a greater impact on resistance profile. 55
A potential concern is that antiviral agents could blunt the immune response to influenza, leaving treated cases susceptible. 85 , 104 Recurrent influenza infection has been reported in one small paediatric case series, 105 but there is no consistent evidence from adult studies to give cause for wider concern. 106 , 107 Re‐infection has also been observed in untreated children, possibly because of immunological naivety or immaturity. 108 Furthermore, adult clinical trials of antiviral agents consistently show that virologically confirmed cases are prevented more effectively than serologically confirmed cases, 89 , 106 , 109 indicating sufficient exposure for seroconversion without illness.
Pandemic vaccines
Inactivated split virion vaccines based on recent H5N1 isolates from humans are currently under phase I trial. As H5 is novel to humans, high concentrations of antigen, 110 , 111 incorporation of adjuvants 112 and booster doses 110 , 111 , 112 have been necessary to achieve immunogenicity. Some monovalent vaccines against novel HA antigens have maximized yield by using reactogenic whole virus formulations. 113 , 114 Candidate H5 vaccines have afforded protection in neutralization assays 115 and animal models against variant H5 viruses. 116 , 117 Such ‘best guess’ vaccines can be stockpiled for possible use in priming critical subpopulations such as front line healthcare workers. 115 Nevertheless, if a pandemic occurs there will likely be substantial delays in production and supply shortages of strain‐specific vaccines due to limited global manufacturing capacity. 118
There is uncertainty about the best correlates of protection for interpandemic influenza vaccines, 119 let alone for a novel strain. Early observations showed that high titre (1:30–1:40) HI antibody is predictive of clinical protection, 17 , 120 , 121 although this measure is subject to considerable inter‐laboratory variation. 122 For licensure in the EU, The Committee for Proprietary Medicinal Products (CPMP) requires documentation of HI antibody responses in terms of: post vaccine seroprotection rate (HI titre ≥1:40), mean fold increase and response, and seroconversion rate. 123 Interpretation is complicated, as fold rises will be smaller for individuals with pre‐existing immunity, although protection will often be improved. 124 Reduced protection against heterologous influenza strains has been shown in challenge studies, 125 , 126 which could explain why some immunogenic vaccines perform poorly in the field. 127 Heterosubtypic vaccines providing broad protection against different sub‐types offer theoretical advantages, but their efficacy is yet to be demonstrated. 128
Strategic vaccine use
An efficient use of pandemic influenza vaccines would be to control transmission through herd immunity. Immunization of pre‐school children in day care is known to significantly protect their families. 129 A more modest reduction in respiratory infections is seen among parents of vaccinated school age children, 130 and in teachers and classmates where institutional coverage is high. 131 In Tecumseh, Michigan, vaccination of schoolchildren prior to the influenza season in 1968–1969 prevented an epidemic similar to that in the comparison city of Adrian. 132 More dramatically, when mandatory influenza vaccination of Japanese schoolchildren was stopped in 1994 there was a sharp increase in pneumonia and influenza mortality in the elderly. 133 Likewise, immunization of carers in aged care institutions predicts a reduction in all‐cause mortality among elderly residents in the UK 66 , 134 and Japan. 67
In the simplest models, immunization moves all vaccinated susceptibles to the removed/recovered class. 72 However, vaccination is more likely to provide incomplete protection for most of those immunized. This critical distinction determines not only the threshold size of an epidemic, 135 but the optimal vaccine coverage required for disease control. 136 Vaccine effects can be characterized further within subgroups of interest identified from clinical trials. 137 In addition, the time to maximal immune response can be a critical consideration in outbreak settings. 36 As most models assume that vaccination reduces both acquisition rate in those exposed, and viral shedding in established cases, it follows that vaccines will be useful even when efficacy is partial. 35 , 37 , 63 , 138
Several models have explored the theoretical effectiveness of targeted immunization for pandemic control, showing optimal benefit from vaccinating children of school 63 , 138 and preschool age. 137 Britton and Becker concluded that influenza could be controlled most efficiently by targeting immunization to families with three or more members. 139 Such model conclusions depend upon underlying assumptions about population susceptibility and transmission in heterogeneous circumstances. For example, using assumptions based on age‐specific attack profiles from the 1957 and 1968 pandemics, Patel et al. obtained two very different estimates for the optimal proportional distribution by age group of a limited number of vaccine doses. 140
Conclusions
Historical and contemporary data show that the transmissibility and pathogenicity of influenza viruses depend on complex interactions with host populations. The emergence of H5N1 as a threat to the poultry industry has undoubtedly been influenced by a very rapid increase in numbers of caged birds in developing countries. Likewise, the risk of a new human pandemic can be linked to both the emergence of avian influenza, and to the large numbers of people on the planet, any one of whom could be host to viruses undergoing a pandemic mutation or recombination event.
Host immunity, which plays a crucial role, has clear specificity for influenza sub‐type and strain. However, cross‐reactive immune responses are needed to explain some otherwise anomalous observations. 21 , 22 In particular, there is reason to believe that clinical attack rates for influenza are not often constrained by low values for R 0, but rather by pre‐existing immunity, and by subclinical infections which can immunize without causing symptoms.
With such complex interactions, making precise predictions about age‐specific attack rates, morbidity and mortality due to a novel pandemic strain is near impossible. Nevertheless, mathematical models do provide a useful framework for pandemic scenarios to explore the potential benefit of public health interventions. As all models have to oversimplify complex biological systems, they should be interpreted with great caution. In particular, models are exquisitely sensitive to underlying assumptions about susceptibility, subpopulations and modes of transmission – all of which must be inferred, rather than being quantified by direct observation.
Model assumptions and predictions should be tested against as many real‐world observations as possible to test the sensitivity of the model to different contexts. By validating model outputs against multiple data sources, investigators can tease out which of the parameters determining disease spread are more likely to vary between contexts, and which are relatively constant. To encourage such work, and to make relevant data sets more easily accessible, we have developed an on‐line publicly searchable archive (FluWeb: http://influenza.sph.unimelb.edu.au), which includes rare historical documents from the 1889–1891 and 1918–1919 pandemics giving individual and group level data on influenza morbidity and mortality. By maximizing our use of historical evidence, we hope to more confidently predict the future.
Conflict of interest
J.D.M. is an active member of the National Influenza Pandemic Advisory Committee (NIPAC). He has advised the Australian Government on influenza and other public health matters as an employee (1999–2004) or occasional consultant (2004–2006) and in these roles has access to information restricted by confidentiality considerations. He has no involvement with professional organizations that may benefit financially from the conclusions of this review.
Acknowledgements
We thank the National Health & Medical Research Council of Australia for funding the database project and associated modelling work (Grant Nos 400588 and 454645). Jodie McVernon is supported by an NHMRC Australian Training Research Fellowship (No. 359238). We thank Terry Nolan, James McCaw, Lorena Brown and other colleagues for helpful comments on the draft manuscript.
References
- 1. Monto AS. The threat of an avian influenza pandemic. N Engl J Med 2005;352:323–325. [DOI] [PubMed] [Google Scholar]
- 2. Hatta M, Kawaoka Y. The continued pandemic threat posed by avian influenza viruses in Hong Kong. Trends Microbiol 2002;10:340–344. [DOI] [PubMed] [Google Scholar]
- 3. Ducatez MF, Olinger CM, Owoade AA et al. Avian flu: multiple introductions of H5N1 in Nigeria. Nature 2006;442:37. [DOI] [PubMed] [Google Scholar]
- 4. Update: WHO‐confirmed human cases of avian influenza A (H5N1) infection, 25 November 2003–24 November 2006. Wkly Epidemiol Rec 2007;82:41–47. [PubMed] [Google Scholar]
- 5. Katz JM, Lim W, Bridges CB et al. Antibody response in individuals infected with avian influenza A (H5N1) viruses and detection of anti‐H5 antibody among household and social contacts. J Infect Dis 1999;180:1763–1770. [DOI] [PubMed] [Google Scholar]
- 6. Butler D. Family tragedy spotlights flu mutations. Nature 2006;442:114–115. [DOI] [PubMed] [Google Scholar]
- 7. Parsons D. Report on the Influenza Epidemic of 1889–90 [monograph on the Internet]. London, England: Local Government Board, Her Majesty's Stationery Office, 1891. FluWeb (Source ID 19): http://influenza.sph.unimelb.edu.au [accessed 31 August 2006]. [Google Scholar]
- 8. Coker R, Mounier‐Jack S. Pandemic influenza preparedness in the Asia‐Pacific region. Lancet 2006;368:886–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mills CE, Robins JM, Lipsitch M. Transmissibility of 1918 pandemic influenza. Nature 2004;432:904–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferguson NM, Cummings DAT, Cauchemez S et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature 2005;437:209–214. [DOI] [PubMed] [Google Scholar]
- 11. Ferguson NM, Cummings DAT, Fraser C, Cajka J, Cooley P, Burke D. Strategies for mitigating an influenza pandemic. Nature 2006;442:448–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003;362:1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 1983;127:361–373. [DOI] [PubMed] [Google Scholar]
- 14. Hayden FG, Croisier A. Transmission of avian influenza viruses to and between humans. J Infect Dis 2005;192:1311–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito T, Couceiro JN, Kelm S et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 1998;72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kash J, Tumpey TM, Proll S et al. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 2006;443:578–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hobson D, Curry R, Beare AS, Ward‐Gardner A. The role of serum haemagglutination‐inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond) 1972;70:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Crosby A. Chapter 12. Samoa and Alaska, in America's Forgotten Pandemic. The Influenza of 1918. Cambridge, UK: Cambridge University Press, 2003. [Google Scholar]
- 19. Ministry of Health . Pandemic of Influenza 1918–9. Reports on Public Health and Medical Subjects No. 4. [monograph on the Internet]. London, UK: His Majesty's Stationery Office, 1920. FluWeb (Source ID 1): http://influenza.sph.unimelb.edu.au [accessed 30 August 2006]. [Google Scholar]
- 20. Mantle J, Tyrrell DA. An epidemic of influenza on Tristan da Cunha. J Hyg (Lond) 1973;71:89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jordan WS Jr, Denny FW Jr, Badger GF et al. A study of illness in a group of Cleveland families. XVII. The occurrence of Asian influenza. Am J Hyg 1958;68:190–212. [DOI] [PubMed] [Google Scholar]
- 22. Epstein SL. Prior H1N1 influenza infection and susceptibility of Cleveland Family Study participants during the H2N2 pandemic of 1957: an experiment of nature. J Infect Dis 2006;193:49–53. [DOI] [PubMed] [Google Scholar]
- 23. Davis L, Caldwell G, Lynch R, Bailey R, Chin T. Hong Kong influenza: the epidemiologic features of a high school family study analyzed and compared with a similar study during the 1957 Asian influenza epidemic. Am J Epidemiol 1970;92:240–247. [DOI] [PubMed] [Google Scholar]
- 24. Hope‐Simpson RE. First outbreak of Hong Kong influenza in a general practice population in Great Britain. A field and laboratory study. Br Med J 1970;3:74–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hall CE, Cooney MK, Fox JP. The Seattle virus watch. IV. Comparative epidemiologic observations of infections with influenza A and B viruses, 1965–1969, in families with young children. Am J Epidemiol 1973;98:365–380. [DOI] [PubMed] [Google Scholar]
- 26. Zachary IG, Johnson KM. Hong Kong influenza in the Panama Canal Zone. First epidemic by a new variant in the western hemisphere. Am J Trop Med Hyg 1969;18:1048–1056. [DOI] [PubMed] [Google Scholar]
- 27. Taber LH, Paredes A, Glezen WP, Couch RB. Infection with influenza A/Victoria virus in Houston families, 1976. J Hyg (Lond) 1981;86:303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fox JP, Hall CE, Cooney MK, Foy HM. Influenzavirus infections in Seattle families, 1975–1979. I. Study design, methods and the occurrence of infections by time and age. Am J Epidemiol 1982;116:212–227. [DOI] [PubMed] [Google Scholar]
- 29. Fox JP, Cooney MK, Hall CE, Foy HM. Influenzavirus infections in Seattle families, 1975–1979. II. Pattern of infection in invaded households and relation of age and prior antibody to occurrence of infection and related illness. Am J Epidemiol 1982;116:228–242. [DOI] [PubMed] [Google Scholar]
- 30. Smirnov YA, Gitelman AK, Govorkova EA, Lipatov AS, Kaverin NV. Influenza H5 virus escape mutants: immune protection and antibody production in mice. Virus Res 2004;99:205–208. [DOI] [PubMed] [Google Scholar]
- 31. Moss P. Cellular immune responses to influenza. Dev Biol (Basel) 2003;115:31–37. [PubMed] [Google Scholar]
- 32. Kreijtz JH, Bodewes R, Van Amerongen G et al. Primary influenza A virus infection induces cross‐protective immunity against a lethal infection with a heterosubtypic virus strain in mice. Vaccine 2007;25:612–620. [DOI] [PubMed] [Google Scholar]
- 33. Cox R, Haaheim L, Ericsson J‐C, Madhun A, Brokstad K. The humoral and cellular responses induced locally and systemically after parenteral influenza vaccination in man. Vaccine 2006;24:6577–6580. [DOI] [PubMed] [Google Scholar]
- 34. Spicer CC, Lawrence CJ. Epidemic influenza in Greater London. J Hyg (Lond) 1984;93:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Longini IM Jr, Nizam A, Xu S et al. Containing pandemic influenza at the source. Science 2005;309:1083–1087. [DOI] [PubMed] [Google Scholar]
- 36. Elveback LR, Fox JP, Ackerman E, Langworthy A, Boyd M, Gatewood L. An influenza simulation model for immunization studies. Am J Epidemiol 1976;103:152–165. [DOI] [PubMed] [Google Scholar]
- 37. Ferguson NM, Mallett S, Jackson H, Roberts N, Ward P. A population‐dynamic model for evaluating the potential spread of resistant influenza virus infections during community‐based use of antivirals. J Antimicrob Chemother 2003;51:977–990. [DOI] [PubMed] [Google Scholar]
- 38. Longini IM Jr, Koopman JS, Haber M, Cotsonis GA. Statistical inference for infectious diseases. Risk‐specific household and community transmission parameters. Am J Epidemiol 1988;128:845–859. [DOI] [PubMed] [Google Scholar]
- 39. Dudley S. The Spread of ‘Droplet Infection’ in Semi‐isolated Communities. Medical Research Council Special Report Series No. 111. [monograph on the Internet]. Oxford, UK: His Majesty's Stationery Office, 1926. FluWeb (Source ID 3): http://influenza.sph.unimelb.edu.au [accessed 13 October 2006]. [Google Scholar]
- 40. Al‐Nakib W, Higgins PG, Willman J et al. Prevention and treatment of experimental influenza A virus infection in volunteers with a new antiviral ICI 130,685. J Antimicrob Chemother 1986;18:119–129. [DOI] [PubMed] [Google Scholar]
- 41. Hayden FG, Fritz R, Lobo MC, Alvord W, Strober W, Straus SE. Local and systemic cytokine responses during experimental human influenza A virus infection. Relation to symptom formation and host defense. J Clin Invest 1998;101:643–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hall CB, Dolin R, Gala CL et al. Children with influenza A infection: treatment with rimantadine. Pediatrics 1987;80:275–282. [PubMed] [Google Scholar]
- 43. Frank AL, Taber LH, Wells CR, Wells JM, Glezen WP, Paredes A. Patterns of shedding of myxoviruses and paramyxoviruses in children. J Infect Dis 1981;144:433–441. [DOI] [PubMed] [Google Scholar]
- 44. Hayden FG. Prevention and treatment of influenza in immunocompromised patients. Am J Med 1997;102(3A):55–60. [DOI] [PubMed] [Google Scholar]
- 45. Mutsch M, Tavernini M, Marx A et al. Influenza virus infection in travelers to tropical and subtropical countries. Clin Infect Dis 2005;40:1282–1287. [DOI] [PubMed] [Google Scholar]
- 46. Moser MR, Bender TR, Margolis HS, Noble GR, Kendal AP, Ritter DG. An outbreak of influenza aboard a commercial airliner. Am J Epidemiol 1979;110:1–6. [DOI] [PubMed] [Google Scholar]
- 47. Lloyd‐Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature 2005;438:355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Isaacs A, Negroni G, Tyrrell DA. Infection of volunteers with Asian influenza virus. Lancet 1957;273:886–887. [DOI] [PubMed] [Google Scholar]
- 49. Herlocher ML, Truscon R, Elias S et al. Influenza viruses resistant to the antiviral drug oseltamivir: transmission studies in ferrets. J Infect Dis 2004;190:1627–1630. [DOI] [PubMed] [Google Scholar]
- 50. Dushoff J, Plotkin JB, Levin SA, Earn DJ. Dynamical resonance can account for seasonality of influenza epidemics. Proc Natl Acad Sci U S A 2004;101:16915–16916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oxford JS, Lambkin R, Sefton A et al. A hypothesis: the conjunction of soldiers, gas, pigs, ducks, geese and horses in northern France during the Great War provided the conditions for the emergence of the ‘‘Spanish’’ influenza pandemic of 1918–1919. Vaccine 2005;23:940–945. [DOI] [PubMed] [Google Scholar]
- 52. Viboud C, Boelle PY, Cauchemez S et al. Risk factors of influenza transmission in households. Br J Gen Pract 2004;54:684–689. [PMC free article] [PubMed] [Google Scholar]
- 53. Cauchemez S, Carrat F, Viboud C, Valleron AJ, Boelle PY. A Bayesian MCMC approach to study transmission of influenza: application to household longitudinal data. Stat Med 2004;23:3469–3487. [DOI] [PubMed] [Google Scholar]
- 54. Longini IM Jr, Monto AS, Koopman JS. Statistical procedures for estimating the community probability of illness in family studies: rhinovirus and influenza. Int J Epidemiol 1984;13:99–106. [DOI] [PubMed] [Google Scholar]
- 55. Stilianakis N, Perelson A, Hayden FG. Emergence of drug resistance during an influenza epidemic: insights from a mathematical model. J Infect Dis 1998;177:863–873. [DOI] [PubMed] [Google Scholar]
- 56. Longini IM Jr, Halloran ME, Nizam A, Yang Y. Containing pandemic influenza with antiviral agents. Am J Epidemiol 2004;159:623–633. [DOI] [PubMed] [Google Scholar]
- 57. Cheung CY, Poon LL, Lau AS et al. Induction of proinflammatory cytokines in human macrophages by influenza A (H5N1) viruses: a mechanism for the unusual severity of human disease? Lancet 2002;360:1831–1837. [DOI] [PubMed] [Google Scholar]
- 58. Lipatov AS, Andreansky S, Webby RJ et al. Pathogenesis of Hong Kong H5N1 influenza virus NS gene reassortants in mice: the role of cytokines and B‐ and T‐cell responses. J Gen Virol 2005;86(Pt 4):1121–1130. [DOI] [PubMed] [Google Scholar]
- 59. Fraser C, Riley S, Anderson RM, Ferguson NM. Factors that make an infectious disease outbreak controllable. Proc Natl Acad Sci USA 2004;101:6146–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Armstrong C, Hopkins R. An epidemiologic study of the 1920 epidemic of influenza in an isolated rural community Public Health Reports No. 36, July 22 [monograph on the Internet], 1921. FluWeb (Source ID 7): http://influenza.sph.unimelb.edu.au [accessed 30 August 2006]. [Google Scholar]
- 61. Foy HM, Cooney MK, Hall CB, Malmgren J, Fox JP. Case‐to‐case intervals of Rhinovirus and Influenza virus infections in households. J Infect Dis 1988;157:180–182. [DOI] [PubMed] [Google Scholar]
- 62. Longini IM Jr, Seaholm SK, Ackerman E, Koopman JS, Monto AS. Simulation studies of influenza epidemics: assessment of parameter estimation and sensitivity. Int J Epidemiol 1984;13:496–501. [DOI] [PubMed] [Google Scholar]
- 63. Germann TC, Kadau K, Longini IM Jr, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 2006;103:5935–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vaughan W. Influenza: An Epidemiological Study. The American Journal of Hygiene, Monographic Series No. 1 [monograph on the Internet]. Lancaster, USA: The New Era Printing Company, 1921. FluWeb (Source ID 2): http://influenza.sph.unimelb.edu.au [accessed 30 August 2006]. [Google Scholar]
- 65. Centre for Communicable Disease Surveillance . Influenza in a boarding school. BMJ 1978. [Google Scholar]
- 66. Potter J, Stott DJ, Roberts MA et al. Influenza vaccination of health care workers in long‐term‐care hospitals reduces the mortality of elderly patients. J Infect Dis 1997;175:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Oshitani H, Saito R, Seki N et al. Influenza vaccination levels and influenza‐like illness in long‐term‐care facilities for elderly people in Niigata, Japan, during an influenza A (H3N2) epidemic. Infect Control Hosp Epidemiol 2000;21:728–730. [DOI] [PubMed] [Google Scholar]
- 68. McCracken K, Curson P. Flu downunder: a demographic and geographic analysis of the 1919 epidemic in Sydney, Australia; in Phillips H, Killingray D. (eds): The Spanish Influenza Pandemic of 1918–19. New Perspectives. London: Routledge, 2003. [Google Scholar]
- 69. Anderson RM, May R. Age‐related changes in the rate of disease transmission: implications for the design of vaccination programmes. J Hyg (Lond) 1985;94:365–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Longini IM Jr, Koopman JS, Monto AS, Fox JP. Estimating household and community transmission parameters for influenza. Am J Epidemiol 1982;115:736–751. [DOI] [PubMed] [Google Scholar]
- 71. Saramaki J, Kaski K. Modelling development of epidemics with dynamic small‐world networks. J Theor Biol 2005;234:413–421. [DOI] [PubMed] [Google Scholar]
- 72. Keeling M. The effects of local spatial structure on epidemiological invasions. Proc Biol Sci 1999; p. 859–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hagenaars TJ, Donnelly CA, Ferguson NM. Spatial heterogeneity and the persistence of infectious diseases. J Theor Biol 2004;229:349–359. [DOI] [PubMed] [Google Scholar]
- 74. Nasell I. On the time to extinction in recurrent epidemics. J R Stat Soc Ser B Stat Method 1999;61:309–330. [Google Scholar]
- 75. Sattenspiel L, Mobarry A, Herring DA. Modeling the influence of settlement structure on the spread of influenza among communities. Am J Hum Biol 2000;12:736–748. [DOI] [PubMed] [Google Scholar]
- 76. Longini IM Jr, Fine PE, Thacker SB. Predicting the global spread of new infectious agents. Am J Epidemiol 1986;123:383–391. [DOI] [PubMed] [Google Scholar]
- 77. Pitman RJ, Cooper BS, Trotter CL, Gay NJ, Edmunds WJ. Entry screening for severe acute respiratory syndrome (SARS) or influenza: policy evaluation. BMJ 2005;331:1242–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heymann A, Chodick G, Reichman B, Kokia E, Laufer J. Influence of school closure on the incidence of viral respiratory diseases among children and on health care utilization. Pediatr Infect Dis J 2004;23:675–677. [DOI] [PubMed] [Google Scholar]
- 79. Becker NG, Glass K, Li Z, Aldis G. Controlling emerging infectious diseases like SARS. Math Biosci 2005;193:205–221. [DOI] [PubMed] [Google Scholar]
- 80. Loeb M, McGeer A, Henry B et al. SARS among critical care nurses, Toronto. Emerg Infect Dis 2004;10:251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yen MY, Lin YE, Su IJ et al. Using an integrated infection control strategy during outbreak control to minimize nosocomial infection of severe acute respiratory syndrome among healthcare workers. J Hosp Infect 2006;62:195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Muller M, McGeer A. Febrile respiratory illness in the intensive care unit setting: an infection control perspective. Curr Opin Crit Care 2006;12:37–42. [DOI] [PubMed] [Google Scholar]
- 83. Tang J, Li Y, Eames I, Chan P, Ridgway G. Factors involved in the aerosol transmission of infection and control of ventilation in healthcare premises. J Hosp Infect 2006;64:100–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Turner D, Wailoo A, Nicholson K, Cooper N, Sutton A, Abrams K. Systematic review and economic decision modelling for the prevention and treatment of influenza A and B. Health Technol Assess 2003;7:1–170. [DOI] [PubMed] [Google Scholar]
- 85. Jefferson T, Demicheli V, Rivetti D, Jones M, Di Pietrantonj C, Rivetti A. Antivirals for influenza in healthy adults: systematic review. Lancet 2006;367:303–313. [DOI] [PubMed] [Google Scholar]
- 86. Boivin G, Coulombe Z, Wat C. Quantification of the influenza virus load by real‐time polymerase chain reaction in nasopharyngeal swabs of patients treated with oseltamivir. J Infect Dis 2003;188:578–580. [DOI] [PubMed] [Google Scholar]
- 87. Writing Committee of the World Health Organization Consultation on Human Influenza A/H5. A/H5 WCotWHOCoHI. Avian influenza A (H5N1) infection in humans. N Engl J Med 2005;353:1374–1384. [DOI] [PubMed] [Google Scholar]
- 88. Nicholson KG, Aoki FY, Osterhaus AD et al. Efficacy and safety of oseltamivir in treatment of acute influenza: a randomised controlled trial. Neuraminidase Inhibitor Flu Treatment Investigator Group. Lancet 2000;355:1845–1850. [DOI] [PubMed] [Google Scholar]
- 89. Hayden FG, Treanor JJ, Fritz RS et al. Use of the oral neuraminidase inhibitor oseltamivir in experimental human influenza: randomized controlled trials for prevention and treatment. JAMA 1999;282:1240–1246. [DOI] [PubMed] [Google Scholar]
- 90. Whitley RJ, Hayden FG, Reisinger KS et al. Oral oseltamivir treatment of influenza in children. Pediatr Infect Dis J 2001;20:127–133. [DOI] [PubMed] [Google Scholar]
- 91. Cooper NJ, Sutton AJ, Abrams KR, Wailoo A, Turner D, Nicholson KG. Effectiveness of neuraminidase inhibitors in treatment and prevention of influenza A and B: systematic review and meta‐analyses of randomised controlled trials. BMJ 2003;326:1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Matheson N, Harnden A, Perera R, Sheikh A, Symmonds‐Abrahams M. Neuraminidase inhibitors for preventing and treating influenza in children. Cochrane Database Syst Rev 2007:CD002744. [DOI] [PubMed] [Google Scholar]
- 93. Hayden FG, Belshe R, Villanueva C et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis 2004;189:440–449. [DOI] [PubMed] [Google Scholar]
- 94. McCaw J, McVernon J. Prophylaxis or treatment? Optimal use of an antiviral stockpile during an influenza pandemic. Math Biosci 2007; Mar 12; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 95. Zambon M, Hayden FG. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res 2001;49:147–156. [DOI] [PubMed] [Google Scholar]
- 96. Monto AS, McKimm‐Breschkin JL, Macken C et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 2006;50:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kiso M, Mitamura K, Sakai‐Tagawa Y et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 2004;364:759–765. [DOI] [PubMed] [Google Scholar]
- 98. Jackson D, Barclay W, Zurcher T. Characterization of recombinant influenza B viruses with key neuraminidase inhibitor resistance mutations. J Antimicrob Chemother 2005;55:162–169. [DOI] [PubMed] [Google Scholar]
- 99. Herlocher ML, Carr J, Ives J et al. Influenza virus carrying an R292K mutation in the neuraminidase gene is not transmitted in ferrets. Antiviral Res 2002;54:99–111. [DOI] [PubMed] [Google Scholar]
- 100. Moscona A. Oseltamivir‐resistant influenza? Lancet 2004;364:733–734. [DOI] [PubMed] [Google Scholar]
- 101. Ferguson NM, Galvani A, Bush R. Ecological and immunological determinants of influenza evolution. Nature 2003;422:428–433. [DOI] [PubMed] [Google Scholar]
- 102. Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 1998;178:1257–1262. [DOI] [PubMed] [Google Scholar]
- 103. Zurcher T, Yates PJ, Daly J et al. Mutations conferring zanamivir resistance in human influenza virus N2 neuraminidases compromise virus fitness and are not stably maintained in vitro. J Antimicrob Chemother 2006;58:723–732. [DOI] [PubMed] [Google Scholar]
- 104. Oker‐Blom N, Hovi T, Leinikki P, Palosuo T, Pettersson R, Suni J. Protection of man from natural infection with influenza A2 Hong Kong virus by amantadine: a controlled field trial. Br Med J 1970;3:676–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ijichi S, Ijichi N. Too early cure of influenza: recurrence in oseltamivir‐treated children. J Paediatr Child Health 2003;39:480–481. [DOI] [PubMed] [Google Scholar]
- 106. Galbraith AW, Oxford JS, Schild GC, Watson GI. Protective effect of 1‐adamantanamine hydrochloride on influenza A2 infections in the family environment: a controlled double‐blind study. Lancet 1969;2:1026–1028. [DOI] [PubMed] [Google Scholar]
- 107. Peters PH Jr, Gravenstein S, Norwood P et al. Long‐term use of oseltamivir for the prophylaxis of influenza in a vaccinated frail older population. J Am Geriatr Soc 2001;49:1025–1031. [DOI] [PubMed] [Google Scholar]
- 108. Hampson AW. Influenza virus antigens and ‘antigenic drift’; in Potter CW. (ed): Influenza. 1st edn. Amsterdam, Boston: Elsevier, 2002. [Google Scholar]
- 109. Hayden FG, Atmar RL, Schilling M et al. Use of the selective oral neuraminidase inhibitor oseltamivir to prevent influenza. N Engl J Med 1999;341:1336–1343. [DOI] [PubMed] [Google Scholar]
- 110. Treanor JJ, Wilkinson BE, Masseoud F et al. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 2001;19:1732–1737. [DOI] [PubMed] [Google Scholar]
- 111. Treanor J, Campbell J, Zangwill K, Rowe T, Wolff M. Safety and Immunogenicity of an inactivated subvirion Influenza A (H5N1) vaccine. N Engl J Med 2006;354:1343–1351. [DOI] [PubMed] [Google Scholar]
- 112. Bresson J‐L, Perronne C, Launay O et al. Safety and immunogenicity of an inactivated split‐virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet 2006;367:1657–1664. [DOI] [PubMed] [Google Scholar]
- 113. Hehme N, Engelmann H, Kuenzel W, Neumeier E, Saenger R. Immunogenicity of a monovalent, aluminum‐adjuvanted influenza whole virus vaccine for pandemic use. Virus Res 2004;103:163–171. [DOI] [PubMed] [Google Scholar]
- 114. Hehme N, Engelmann H, Kunzel W, Neumeier E, Sanger R. Pandemic preparedness: lessons learnt from H2N2 and H9N2 candidate vaccines. Med Microbiol Immunol (Berl) 2002;191:203–208. [DOI] [PubMed] [Google Scholar]
- 115. Stephenson I, Bugarini R, Nicholson KG et al. Cross‐reactivity to highly pathogenic avian influenza H5N1 viruses after vaccination with nonadjuvanted and MF59‐adjuvanted influenza A/Duck/Singapore/97 (H5N3) vaccine: a potential priming strategy. J Infect Dis 2005;191:1210–1215. [DOI] [PubMed] [Google Scholar]
- 116. Lipatov AS, Webby RJ, Govorkova EA, Krauss S, Webster RG. Efficacy of H5 influenza vaccines produced by reverse genetics in a lethal mouse model. J Infect Dis 2005;191:1216–1220. [DOI] [PubMed] [Google Scholar]
- 117. Govorkova EA, Webby RJ, Humberd J, Seiler J, Webster RG. Immunization with reverse‐genetics‐produced H5N1 influenza vaccine protects ferrets against Homologous and Heterologous challenge. J Infect Dis 2006;194:159–167. [DOI] [PubMed] [Google Scholar]
- 118. Schwartz B, Gellin B. Vaccination strategies for an influenza pandemic. J Infect Dis 2005;191:1207–1209. [DOI] [PubMed] [Google Scholar]
- 119. Lipsitch M. Ethics of rationing the flu vaccine. Science 2005;307:41. [DOI] [PubMed] [Google Scholar]
- 120. Tyrrell DA. Assessment of vaccine efficacy by challenge studies in man. Postgrad Med J 1976;52:379–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Potter CW, Oxford JS. Determinants of immunity to influenza infection in man. Br Med Bull 1979;35:69–75. [DOI] [PubMed] [Google Scholar]
- 122. Wood JM, Gaines‐Das RE, Taylor J, Chakraverty P. Comparison of influenza serological techniques by international collaborative study. Vaccine 1994;12:167–174. [DOI] [PubMed] [Google Scholar]
- 123. Hannoun C, Megas F, Piercy J. Immunogenicity and protective efficacy of influenza vaccination. Virus Res 2004;103:133–138. [DOI] [PubMed] [Google Scholar]
- 124. Beyer WE, Palache AM, Luchters G, Nauta J, Osterhaus AD. Seroprotection rate, mean fold increase, seroconversion rate: which parameter adequately expresses seroresponse to influenza vaccination? Virus Res 2004;103:125–132. [DOI] [PubMed] [Google Scholar]
- 125. Potter CW, Jennings R, Nicholson K, Tyrrell DA, Dickinson KG. Immunity to attenuated influenza virus WRL 105 infection induced by heterologous, inactivated influenza A virus vaccines. J Hyg (Lond) 1977;79:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Larson HE, Tyrrell DA, Bowker CH, Potter CW, Schild GC. Immunity to challenge in volunteers vaccinated with an inactivated current or earlier strain of influenza A(H3N2). J Hyg (Lond) 1978;80:243–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Tyrrell DA, Buckland R, Rubenstein D, Sharpe DM. Vaccination against Hong Kong influenza in Britain, 1968–9. A report to the Medical Research Council Committee on Influenza and other Respiratory Virus Vaccines. J Hyg (Lond) 1970;68:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Tamura S, Tanimoto T, Kurata T. Mechanisms of broad cross‐protection provided by influenza virus infection and their application to vaccines. Jpn J Infect Dis 2005;58:195–207. [PubMed] [Google Scholar]
- 129. Hurwitz ES, Haber M, Chang A et al. Effectiveness of influenza vaccination of day care children in reducing influenza‐related morbidity among household contacts. JAMA 2000;284:1677–1682. [DOI] [PubMed] [Google Scholar]
- 130. Piedra PA, Gaglani MJ, Kozinetz CA et al. Herd immunity in adults against influenza‐related illnesses with use of the trivalent‐live attenuated influenza vaccine (CAIV‐T) in children. Vaccine 2005;23:1540–1548. [DOI] [PubMed] [Google Scholar]
- 131. Rudenko LG, Slepushkin AN, Monto AS et al. Efficacy of live attenuated and inactivated influenza vaccines in schoolchildren and their unvaccinated contacts in Novgorod, Russia. J Infect Dis 1993;168:881–887. [DOI] [PubMed] [Google Scholar]
- 132. Monto AS, Davenport FM, Napier JA, Francis T Jr Modification of an outbreak of influenza in Tecumseh, Michigan by vaccination of schoolchildren. J Infect Dis 1970;122:16–25. [DOI] [PubMed] [Google Scholar]
- 133. Reichert TA, Sugaya N, Fedson DS, Glezen WP, Simonsen L, Tashiro M. The Japanese experience with vaccinating schoolchildren against influenza. N Engl J Med 2001;344:889–896. [DOI] [PubMed] [Google Scholar]
- 134. Carman WF, Elder AG, Wallace LA et al. Effects of influenza vaccination of health‐care workers on mortality of elderly people in long‐term care: a randomised controlled trial. Lancet 2000;355:93–97. [DOI] [PubMed] [Google Scholar]
- 135. Smith P, Rodrigues L, Fine P. Assessment of the protective efficacy of vaccines against common diseases using case‐control and cohort studies. Int J Epidemiol 1984;13:87–93. [DOI] [PubMed] [Google Scholar]
- 136. Ball F, Britton T, Lyne O. Stochastic multitype epidemics in a community of households: estimation and form of optimal vaccination schemes. Math Biosci 2004;191:19–40. [DOI] [PubMed] [Google Scholar]
- 137. Hill AN, Longini IM Jr. The critical vaccination fraction for heterogeneous epidemic models. Math Biosci 2003;181:85–106. [DOI] [PubMed] [Google Scholar]
- 138. Weycker D, Edelsberg J, Halloran ME et al. Population‐wide benefits of routine vaccination of children against influenza. Vaccine 2005;23:1284–1293. [DOI] [PubMed] [Google Scholar]
- 139. Britton T, Becker NG. Estimating the immunity coverage required to prevent epidemics in a community of households. Biostatistics 2000;1:389–402. [DOI] [PubMed] [Google Scholar]
- 140. Patel R, Longini IM Jr, Halloran ME. Finding optimal vaccination strategies for pandemic influenza using genetic algorithms. J Theor Biol 2005;234:201–212. [DOI] [PubMed] [Google Scholar]


