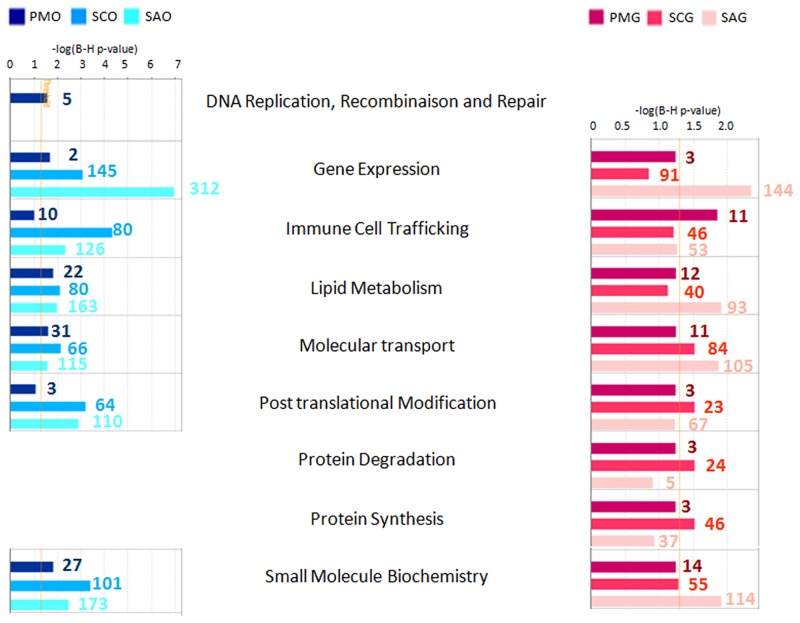
Fig 2. Cellular function enrichment.
Functional enrichment analysis for sets of differentially expressed genes during early follicular development was performed in silico for each compartment using Ingenuity Pathway Analysis (IPA) software. Statistical significance was determined by a P value calculated using Fisher’s exact test corrected for multiple testing correction (Benjamini-Hochberg test, FDR <0.05). This analysis identified nine main cellular function categories as being significantly enriched in differentially expressed genes. Bar colors correspond to enriched functions at primary stage (compared to primordial stage), secondary stage (compared to primary and primordial stages), and small antrum stage (compared to secondary, primary and primordial stages). The X axis corresponds to the level of significance of the function: -log(B-H p value). Granulosa cell functions are colored in red and oocyte functions are colored in blue. Numbers correspond to the number of focus genes contributing to the functions. Follicular stage abbreviations are described in Fig 1.