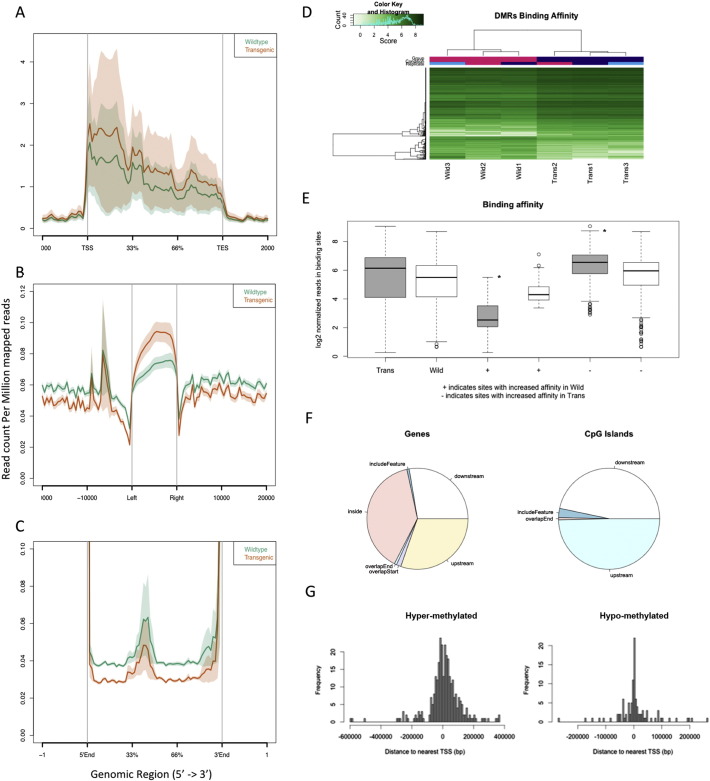
Fig. 4.
DNA methylation patterns in lungs of wild-type and transgenic animals measured by MBD-Seq. (A) Reads of DMR aligned to normalized gene bodies for samples from wild-type (green) and transgenic (brown) animals. The genes were transformed to a body percentage from its length to make the measurements comparable. (B) Overall reads aligned to normalized gene bodies with extended intergenic regions and CpG island shores for samples from wild-type and transgenic animals. (C) Highlight of overall reads aligned to intergenic regions for samples from wild-type and transgenic animals. (D) Clustering of genomic DNA samples from transgenic and wild-type animals based on DMRs. (E) Depiction of DMR methylation for each sample group (wild-type and transgenic). Signs + and − illustrate the regions with incremented affinity in wild-type and transgenic group, respectively. * (p < 0.001 by Wilcoxon test). (F) Summary of the locations of DMRs regarding features, genes and CpG islands. (G) Distribution of DMRs of both hyper-methylated and hypo-methylated in transgenic mice relative to transcription starting site (TSS).