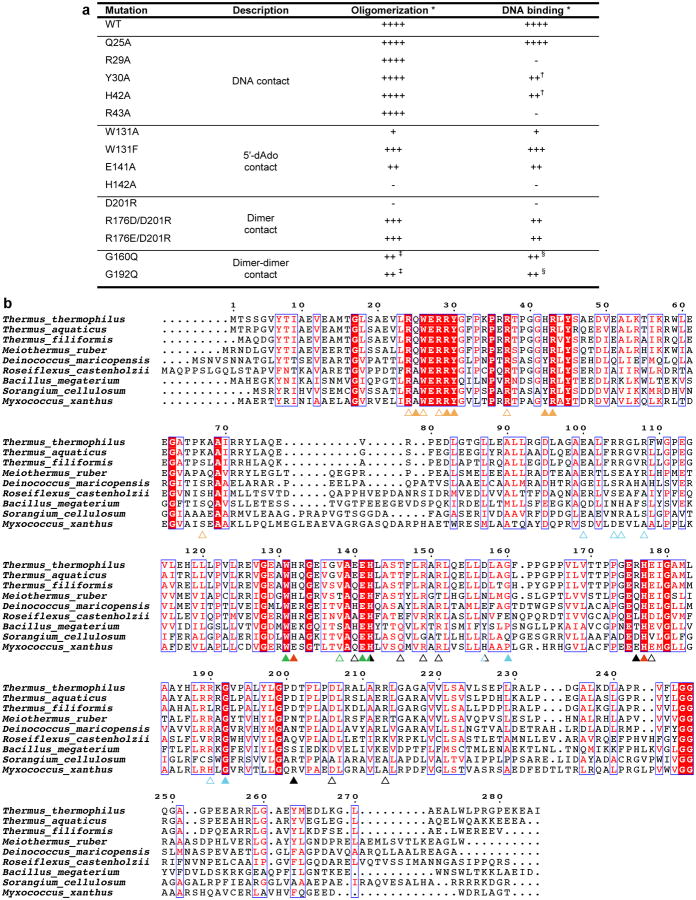
Extended Data Figure 3.
CarH mutant analysis and multiple sequence alignment. (a) Results summary for in vitro CarH mutant analysis. (b) Alignment of CarH sequences from different bacterial species. Sequence identity is shown in white font with red background, sequence similarity in red font. Colored triangles highlight functionally important positions, with filled triangles indicating residues analyzed by mutagenesis in this study and empty triangles indicating residues not analyzed by mutagenesis. Mutating the highly conserved His177 of the Cbl-binding motif, the lower axial ligand of bound AdoCbl, has previously been shown to impair AdoCbl binding and tetramerization8. Coloring as follows: hydrogen bonds/ionic interactions to DNA: orange; contact to 5′-dAdo: green; histidines coordinating Cbl (His132 only coordinates after light exposure): red; hydrogen bonds/ionic interactions at dimer interface: black; hydrogen bonds/ionic interactions as well as Gly160 and Gly192 at dimer-dimer interface: cyan. Residues involved in more than one type of interaction are colored half/half. Residues at protein interfaces are less well conserved than other functionally important residues, likely because compensatory mutations and local structural deformations are possible. Note, however, that the T. thermophilus Arg176-Asp201 pair observed in our structure is inverted in Myxococcus xanthus, suggesting that the interaction is conserved. Alignment generated using ESPript63.