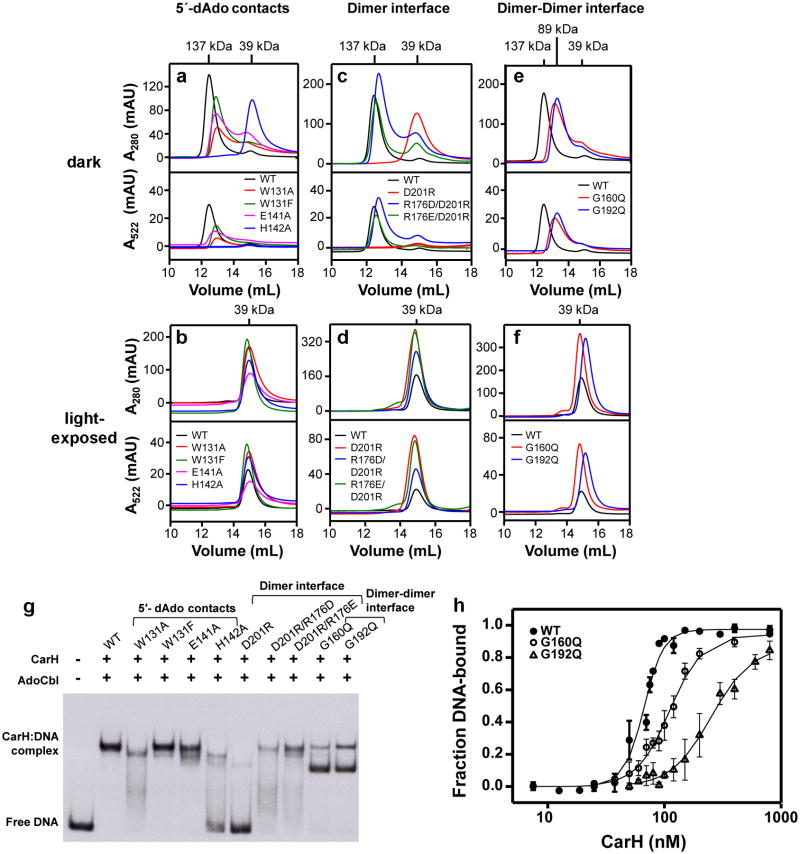
Extended Data Figure 4.
Characterization of CarH mutants affecting oligomerization state. (a-f) SEC traces (Superdex 200 analytical SEC column) of CarH carrying mutations (a,b) near the 5′-dAdo group, (c,d) at the head-to-tail dimer interface, and (e,f) at the dimer-dimer interface. Shown are traces of mutants incubated with AdoCbl in the dark (top panels) and after light exposure (bottom panels). In all panels, both A280 (tracking protein) and A522 (tracking Cbl) traces are shown. Molecular weights are calculated from the observed elution volumes as described in the methods, and are consistent with a tetrameric species (137 kDa), a dimeric species (89 kDa), and a monomeric species (39 kDa). Notably, mutant CarH proteins that do not tetramerize in the presence of AdoCbl (D201R, H142A) also do not appear to bind AdoCbl (see 522 nm traces of dark samples). This finding is consistent with previous studies that show cooperativity of AdoCbl binding and tetramerization, a feature that does not hold for other forms of Cbl (methylcobalamin, CNCbl and Cbl)8. Both of these mutant proteins can still bind Cbl (see 522 nm traces of light-exposed samples), which further suggests that these mutants are properly folded and that the lack of AdoCbl binding stems from inability to oligomerize. Although the degree of tetramerization of CarH mutant proteins in the dark varied, all of these mutant proteins form Cbl-bound monomers after light exposure (g) DNA-binding capacity of WT and mutant proteins (800 nM) as determined by EMSAs after incubation with AdoCbl (4 μM) in the dark. (h) EMSA data for WT CarH and the G160Q and G192Q mutants fit to the Hill equation, as described in the methods. KD (in nM) and Hill coefficients from the fits are, respectively, (67±2) and (5.1±0.7) for WT CarH, (111±18) and (3.0±0.2) for G160Q CarH, and (253±17) and (2.5±0.3) for G192Q CarH. The data shown are the mean values and standard errors of three to five repeat experiments.