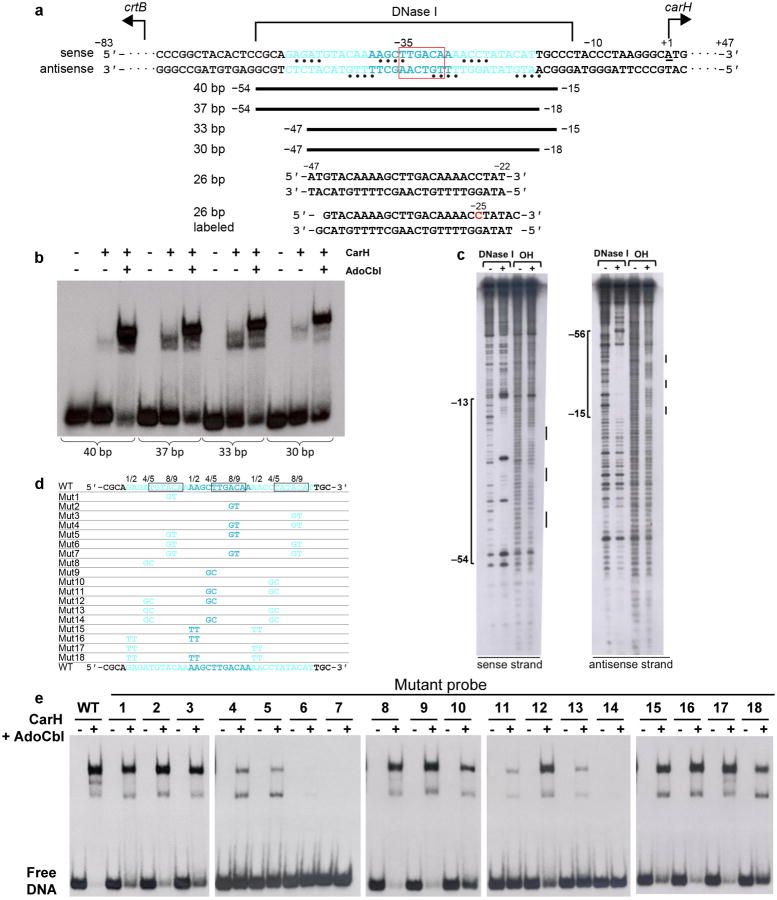
Extended Data Figure 5.
Identification and validation of CarH operator sequence by EMSAs and footprinting. (a) Location of CarH operator in the intergenic region between carH and the carotenogenic crtB of the T. thermophilus genome. Structural and biochemical data are mapped onto the sequence. Three 11-bp CarH binding sites are shown in cyan font and the promoter −35 element is highlighted with a red box. Nucleotides protected from hydroxyl radical cleavage are indicated with bullets. The ∼42-nucleotide DNase I footprint on the sense strand is shown above the sequence and that of the antisense strand has been omitted for clarity. Nucleotide numbering on the sense strand is relative to the carH transcription start site (underlined, +1)10. To identify suitable DNA constructs for crystallization, operator sequences were systematically trimmed around a ∼40-bp segment, as indicated by the black bars, and binding was assessed by EMSAs (shown in b). The sequences of two 26-bp DNA segments used for co-crystallization are also shown. The blunt-ended 26-bp segment was used for determination of the CarH:DNA structure. The second 26-bp segment contained 1-nucleotide 3′-overhangs and 5-iodo-deoxycytidine in position −25 (red) and was used to validate the mode of DNA binding. (b) Binding of CarH (800 nM) to DNA segments of different lengths after incubation with AdoCbl (4 μM) in the dark. Substantial DNA-binding was observed for a probe as small as 30-bp. (c) DNase I and hydroxyl radical footprints of CarH on a 130-bp operator DNA segment. Disappearance of bands in the presence of CarH indicates protection from cleavage. Protected regions are marked on the side and were mapped onto the operator sequence using G+A chemical sequencing experiments performed in parallel. (d,e) CarH binding to 40-bp operators carrying mutations. (d) Sequences of tested operator variants. WT operator sequence shown at the top and bottom, with repeat sequences that CarH recognizes shown in cyan. 6-bp stretch contacted by CarH recognition helix is boxed. Mutations are as follows: Mut1-7: single (1-3), pairwise (4-6), and triple (7) mutations of AC to GT (Positions 8/9); Mut8-14: single (8-10), double (11-13), and triple (14) mutations of (A/C)T to GC (Positions 4/5); Mut15-18: pairwise (15-17) and triple (18) mutations of (A/G)A) to TT (Positions 1/2). (e) EMSAs with WT CarH (800 nM) and each of the 40-bp operator variants after incubation with AdoCbl (4 μM) in the dark. Note that two additional lower mobility complexes are observed, most apparent with the WT operator and its variants with comparable binding. The origin of these complexes is unknown, but they likely arise from oligomeric equilibria and residual amounts of light-exposed protein in the sample.