Fig 3. Spontaneous nuclear and mitochondrial repeat mediated deletions.
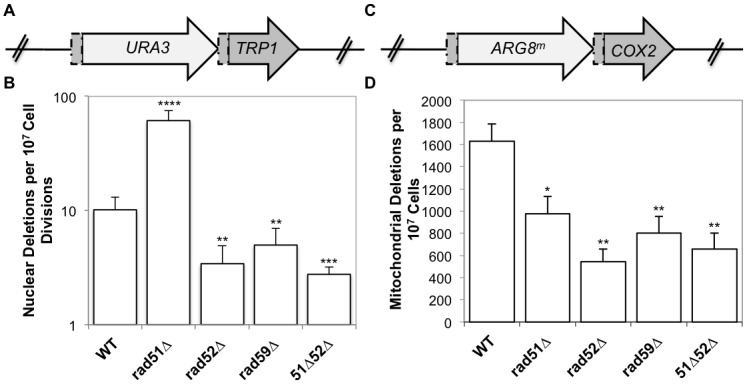
(A) The nuclear DRMD reporter. The URA3 gene was inserted 99 bp into the TRP1 sequence, and is followed directly by the entire TRP1 gene lacking the start codon, resulting in a 96 bp direct repeat flanking the URA3 marker. Spontaneous deletions are selected on medium lacking tryptophan, and rates were determined using the method of the median. (B) The mitochondrial DRMD reporter. The ARG8 m gene was inserted 99 bp into the COX2 sequence, and is followed directly by the entire COX2 gene lacking the start codon, resulting in a 96 bp direct repeat flanking the ARG8 m marker. Spontaneous deletions are selected on medium containing glycerol as the sole carbon source, and rates were determined using the method of the median. (C) Average rates of nuclear DRMD. (D) Average rates of mitochondrial DRMD. Error bars indicate SD. Asterisks indicate significant differences between the mutant and wild-type rates (* = p ≤ 0.05, ** = p ≤ 0.01, *** = p ≤ 0.001, **** = p ≤ 0.0001).
