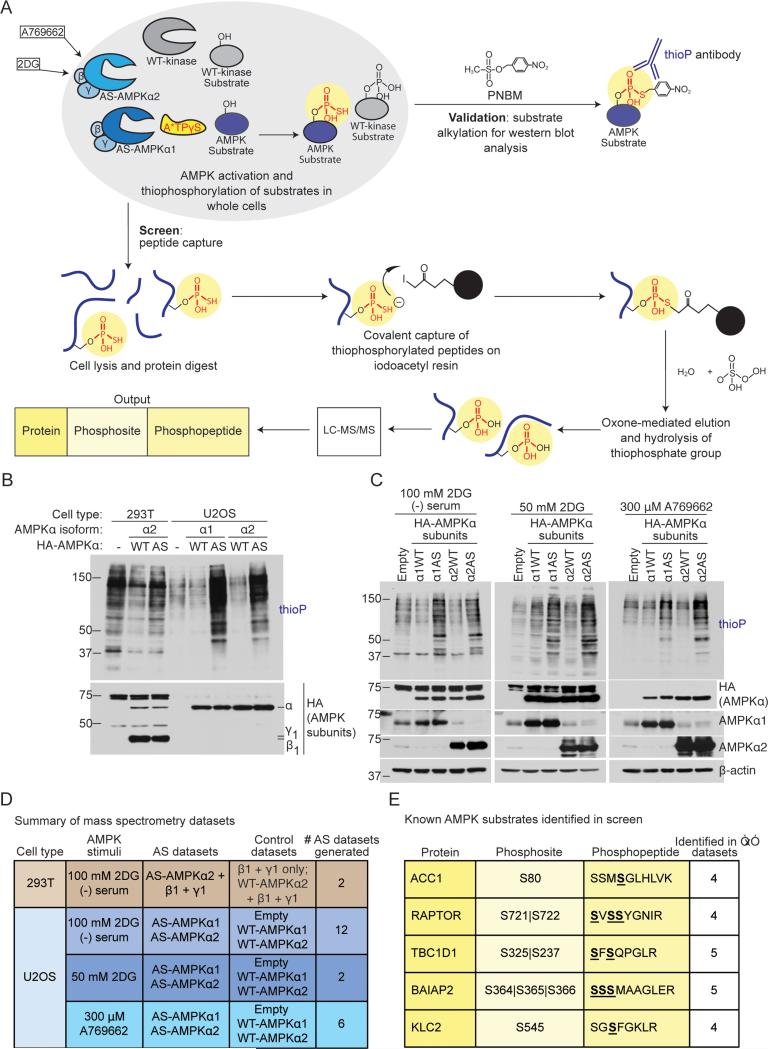
Figure 1. Screening strategy to identify AMPKα1 andα2 substrates and phosphorylation sites in cells.
(A) Schematic of the peptide-capture technique used to identify analog-specific (AS) AMPKα1 and α2 substrates and phosphorylation sites in whole cells. AS-AMPK uses A*TPγS, a bulky ATP analog, to thiophosphorylate substrates. Upper panel: thiophosphorylated substrates are alkylated by p-nitrobenzyl mesylate (PNBM) and recognized by an antibody to the thiophosphate moiety (thioP). Lower panel: thiophosphorylated peptides are captured on a resin, eluted, and identified using liquid chromatography-tandem mass spectrometry (LC-MS/MS). “2DG”, 2-deoxy-D-glucose.
(B) HA-tagged AS-AMPKα1 and α2 thiophosphorylate endogenous substrates in U2OS cells without over-expression of the β and γ subunits. Cells were serum-starved for 2 hours and stimulated for 5 minutes with 100 mM 2DG, then incubated with A*TPγS. Whole cell lysates were analyzed for the presence of thiophosphorylation (thioP) and exogenous AMPK subunits (HA tag).
(C) HA-tagged AS-AMPKα1 and α2 thiophosphorylate endogenous substrates under different AMPK-activating conditions. Whole cell lysates were analyzed for the presence of thiophosphorylation (thioP) and AMPKα (HA tag, AMPKα1, AMPKα2). First panel: 2 hours of serum-starvation with 5 minutes of 100 mM 2DG; second panel: 15 minutes of 50 mM 2DG; third panel: 30 minutes of 300 μM A769662. Representative of 6, 1, and 3 independent experiments for 2DG (−) serum, 2DG, and A769662, respectively. “Empty”, Empty vector; “α1WT”, WT-AMPKα1; “α1AS”, AS-AMPKα1, “α2WT”, WT-AMPKα2, “α2AS”, AS-AMPKα2.
(D) Summary of mass spectrometry datasets. AMPK-activating conditions as in Figure 1C. See Figure S1G and Supplementary List 1 for more information. “Empty”, Empty vector.
(E) Known AMPK substrates identified in multiple AS-AMPK datasets. Underlined and bold residues, phosphorylated sites on the identified phosphopeptide (more than one is shown if the phosphopeptide had multiple or ambiguous phosphorylation site identification). “Phosphosite” column, identified phosphorylation site corresponding to the known AMPK site. “|”, ambiguous site identification. S722, known AMPK site on RAPTOR; S237, known site on TBC1D1; S366, known site on BAIAP2.