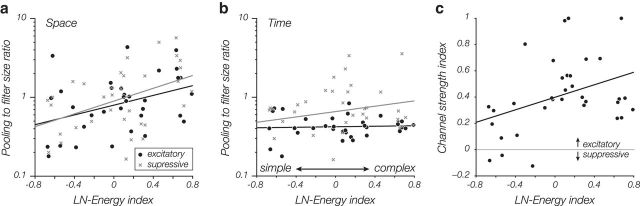
Figure 5.
Dependence of model-estimated pooling extent and strength on cell complexity. The abscissa of each graph represents a complexity index (derived from the LN and energy models as (Eenergy − ELN)/(Eenergy + ELN); see Materials and Methods). A value of −1 indicates a purely simple cell; and a value of 1 indicates a purely complex cell. a, Relative spatial pooling size in the fitted subunit model increases with cell complexity. For each cell, we computed the ratio of SDs of 2D Gaussian envelopes fitted to the pooling weights and the subunit filter. As expected, complex cells pool over a larger relative region than simple cells. This same effect is seen for the suppressive channel (gray x's). b, Relative temporal pooling size is not correlated with cell complexity, but pooling duration of the inhibitory channel is generally larger than that of the excitatory channel. c, Relative strength of excitatory to inhibitory channels increases with cell complexity. Relative strength is computed as the ratio of SDs of the two channel responses (i.e., generator signals) over all stimuli.