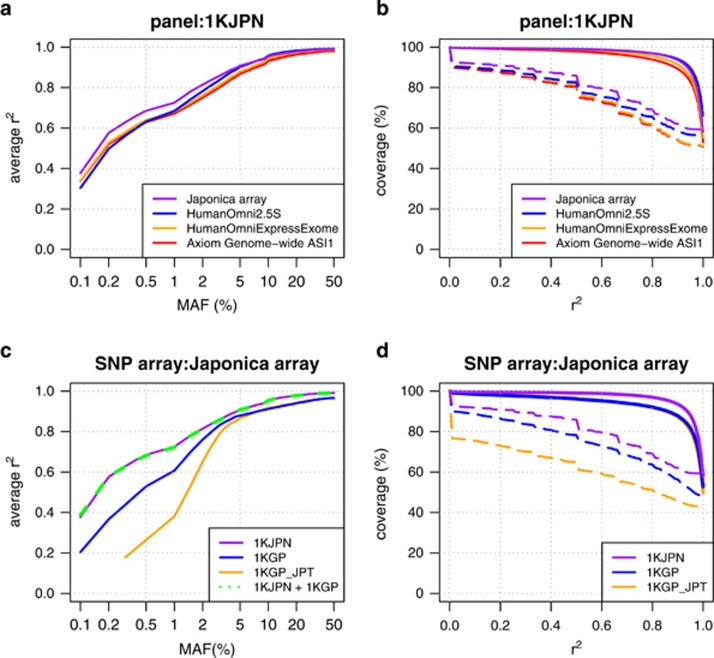
Figure 2.
Improvement in imputation accuracy with the Japonica array. Comparison of the imputation accuracy of different SNP arrays using the 1KJPN panel (a and b) and the imputation accuracy of the Japonica array using different reference panels (c and d). The imputation was conducted to the 131 individuals (ToMMo131, independent from the 1070 individuals in the 1KJPN panel) using the 1KJPN panel. The average r2 values are plotted against the MAF (a and c). The fraction of SNPs in which the genotype was imputed with a given r2 threshold (x-axis) over the total SNPs in the reference panel (genomic coverage) is plotted (b and d) with solid and dashed lines for common and low-frequency SNPs, respectively. The r2 value is the squared correlation coefficient between the imputed genotype and the genotype obtained by whole-genome sequencing. MAF, minor allele frequency; SNP, single nucleotide polymorphism.