Figure 6.
Mapping of the Collagen-Binding Site of Endo180 D1-4
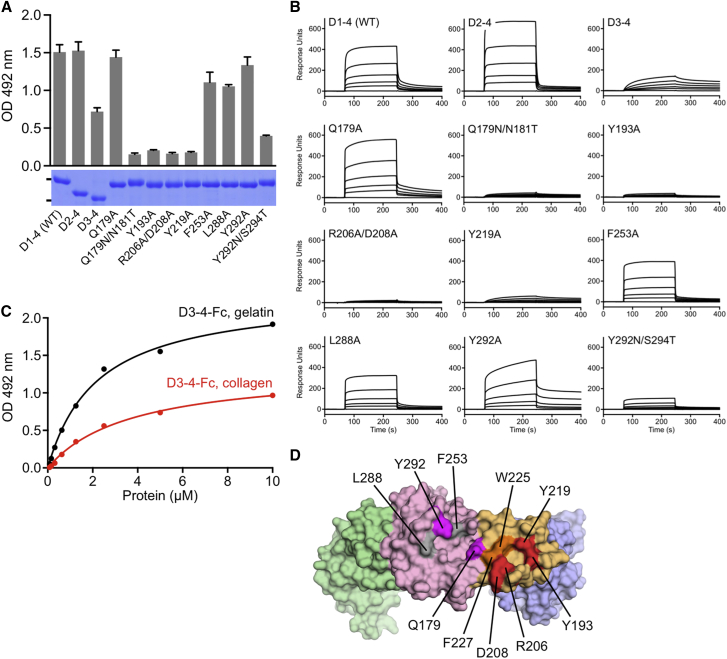
(A) Solid-phase assay of Fc-tagged Endo180 proteins binding to immobilized gelatin. All proteins were tested at a fixed concentration of 1 μM. The data are mean ± SE (n = 3). The Coomassie blue-stained SDS-PAGE gel shows the proteins used in the assay, with the positions of the 100- and 60-kDa markers indicated on the left.
(B) SPR analysis of Fc-tagged Endo180 proteins binding to immobilized gelatin. Raw sensorgrams are shown for five concentrations (2, 1, 0.5, 0.25 and 0.125 μM) and a buffer injection.
(C) Solid-phase assay of Endo180 D3-4-Fc binding to immobilized type I collagen and gelatin. The KD values derived from the fits are 3.7 ± 0.43 and 2.2 ± 0.15 μM, respectively. Endo180 D3-4-Fc binding to an uncoated control surface is negligible.
(D) Point mutations mapped onto the Endo180 D1-4 crystal structure (domains colored as in Figure 1B): red, mutation to alanine reduces collagen binding; magenta, mutation to an N-linked glycosylation site reduces collagen binding; gray, mutation to alanine has no effect on collagen binding; orange, mutation to alanine abolished protein secretion.
See also Figure S4.