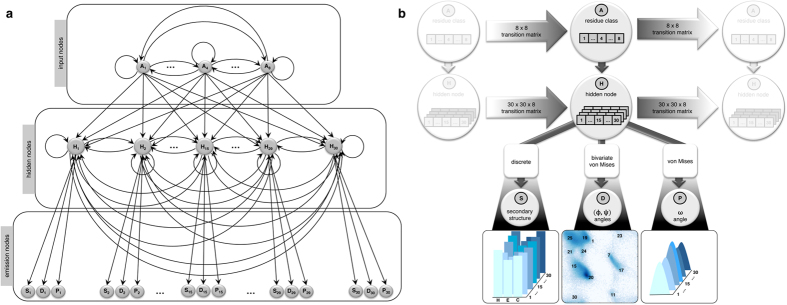
Figure 1. Architecture of FUSION IOHMM.
(a) Circular nodes represent stochastic variables and arrows in the graph specify the conditional independent relationships among variables. Input nodes capture protein’s sequence space while output (i.e., emission) nodes correspond to structural space. Hidden nodes specify dependencies between sequence and structural space along the sequence (i.e., not only between consecutive nodes). (b) In each slice, an input node controls transition of the residue classes in the amino acid sequence (A) and a Markov chain of hidden nodes (H) captures the sequential dependencies along the peptide chain where each hidden node corresponds to three kinds of emission distributions: (1) three-state secondary structure labels (S): helix (H), strand (E), and coil (C), (2) backbone (ϕ, ψ) dihedral angle pairs (D), and (3) ω angles associated with peptide bonds (P). The type of emission distribution is specified in square boxes.