Fig. 3.
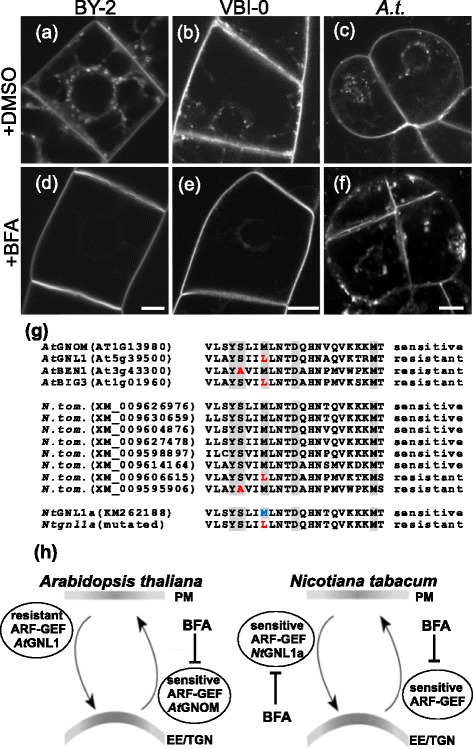
Differential reaction of tobacco and Arabidopsis cells to BFA is reflected by the composition of ARF-GEFs. a-n In vivo confocal microscopy of FM 4–64 (2 μM, 20 min) uptake by 3-day-old tobacco BY-2 and VBI-0 cells and A.t. Col-0 cells pre-treated with mock DMSO or 20 μM BFA for 30 min. Confocal sections of perinuclear plane captured with confocal microscope (561 nm excitation). a-c Control cells with DMSO mock treatment of (a) BY-2 (b) VBI-0 and (c) A.t. FM 4–64 stained endosomes are randomly distributed through the cytoplasm. Note the PM staining. d-f BFA (20 μM)-treated cells. Note the inhibition of FM 4–64 uptake in (d) BY-2 and (e) VBI-0 cells and PM staining. In contrast, (f) Arabidopsis cells show no inhibition of FM 4–64 uptake with the accumulated of signal within the cytoplasm. Note the absence of PM staining. Scale bar = 10 μm. g Amino acid sequence alignment of the region determining BFA sensitivity/resistance of individual ARF-GEFs (Sec7 domain). AtGNOM is an example of a BFA-sensitive ARF-GEF, while AtGNL1, AtBEN1 and AtBIG3 are examples of BFA-resistant ARF-GEFs. Residues known to be involved in BFA sensitivity/resistance are highlighted gray and the ones determining resistance are in red. Eight unique Sec7 domains of potential ARF-GEFs in N. tom. genome are shown. The “BFA sensitivity” determining methionine residue in NtGNL1a, which was modified to leucine is in blue. h Adaptation of the schematic model from Geldner et al. [17] highlighting the role of N.t. BFA-sensitive ARF-GEF (NtGNL1a) in ARF-GEF-dependent endocytosis in contrast to the BFA-resistant ARF-GEF (AtGNL1) in A.t.
