Fig. 5.
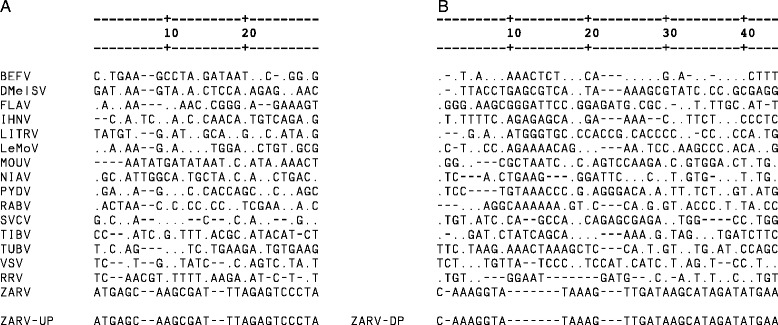
ZARV primer sequence alignment with other representative Rhabdovirus sequences, flanking the putative phosphoprotein/ORF2 gene. a alignment of upstream primer. b alignment of downstream primer. ZARV genome and primer sequences are represented as given. Nucleotides of the other Rhabdovirus sequences represented in the letter code differ from the ZARV sequence. Consereved nucleotides are represented as dots. BEFV: Bovine ephemeral fever virus (KM276084.1), DMelSV: Drosophila melanogaster sigma virus (NC_013135.1), FLAV: Flanders virus (KJ958896.1), IHNV: Infectious hematopoietic necrosis virus (NC_001652.1), LITRV: Long Island tick rhabdovirus (NC_025340.1), LeMoV: Lettuce yellow mottle virus (NC_011532.1), MOUV: Moussa virus (NC_025359.1), NIAV: Niakha virus (NC_025405.1), PYDV: Potato yellow dwarf virus (NC_016136.1), RABV: Rabies virus (AF499686.2), SVCV: Spring viraemia of carp virus (NC_002803.1), TIBV: Tibrogargan virus (GQ294472.1), TUBV: Tupaia virus (NC_007020.1), VSV: Vesicular stomatitis virus (NC_001560.1), PRV: Perch rhabdovirus (NC_020803.1). The alignment was calculated using MegAlign 10.1.0 (DNASTAR) employing the ClustalW algorithm
