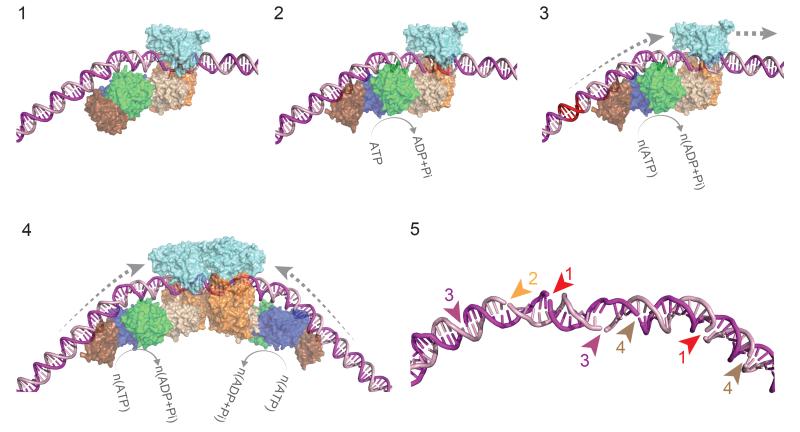
Figure 6. Model for loop-independent DNA translocation and extensive nucleolytic DNA processing.
Model for loop-independent DNA translocation and extensive nucleolytic DNA processing. (1) The pre-initiation complex. The nuclease is in an inactive conformation. (2) The ATPase cycle loosens the MTase-TRD grip on the DNA. (3) dsDNA translocation downstream of the target (red). (4) Convergence of two enzymes initially brings the nucleases approximately 75 bp apart. (5) Example of stochastic “DNA shredding” by a collision complex. Numbers are the order of the nicking events.