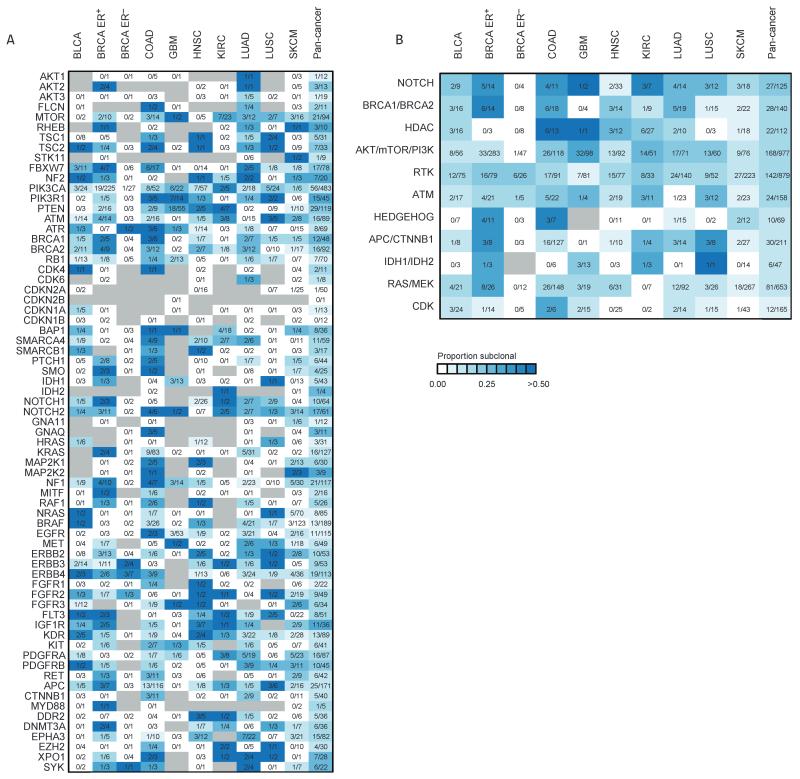
Fig. 4.
Clonal heterogeneity of mutations in genes linked to therapies. (A) Heatmap showing the proportion of nonsilent mutations that are subclonal for each potentially actionable gene across nine cancer types. For each mutation, the number of subclonal mutations identified is indicated for each cancer type and the combined pan-cancer data set. Gray indicates the absence of a mutation. (B) The clonality of actionable pathways is depicted for each cancer type and the combined pan-cancer data set. Pathways are ordered according to subclonality in the pan-cancer data set, with pathways that have a higher proportion of subclonal mutations at the top of the heatmap. Genes related to CDKs (cyclin-dependent kinases) have very few subclonal mutations. RTK, receptor tyrosine kinases. For details of all the genes within each pathway, see table S4.