SUMMARY
The antimicrobial peptide LL-37 is generated upon proteolytic cleavage of cathelicidin and limits invading pathogens by directly targeting microbial membranes as well as stimulating innate immune cell function. However, some microbes evade LL-37-mediated defense. Notably, Group A Streptococcus (GAS) strains belonging to the hypervirulent M1T1 serogroup are more resistant to human LL-37 than other GAS serogroups. We show that the GAS surface-associated M1 protein sequesters and neutralizes LL-37 antimicrobial activity through its N-terminal domain. M1 protein also binds the cathelicidin precursor hCAP-18, preventing its proteolytic maturation into antimicrobial forms. Exogenous M1 protein rescues M1-deficient GAS from killing by neutrophils and within neutrophil extracellular traps and neutralizes LL-37 chemotactic properties. M1 also binds murine cathelicidin, and its virulence contribution in a murine model of necrotizing skin infection is largely driven by its ability to neutralize this host defense peptide. Thus, cathelicidin resistance is essential for the pathogenesis of hyperinvasive M1T1 GAS.
INTRODUCTION
Cathelicidins are cationic antimicrobial peptides produced by leukocytes and epithelial cells that provide a critical first line of defense against microbial invasion (Ho Wong et al., 2013; Nizet et al., 2001). Cathelicidin expression is strongly induced during infection and injury (Dorschner et al., 2001), initially as a full-length protein that lacks antimicrobial activity (Zaiou et al., 2003). Neutrophil proteinase-3 and keratinocyte kallikreins process cathelicidin to liberate antimicrobial peptides derived from its carboxy-terminus (Murakami et al., 2004; Sørensen et al., 2001; Yamasaki et al., 2006). Some cathelicidin peptides, the best characterized being human LL-37, also function as signaling molecules to stimulate inflammation (Yamasaki et al., 2007), production of other antimicrobials (Alalwani et al., 2010), chemotaxis (Yang et al., 2000), and formation of neutrophil extracellular traps (NETs) (Neumann et al., 2014).
Streptococcus pyogenes (group A Streptococcus; GAS) is a leading human pathogen, associated with hundreds of millions of pharyngeal and skin infections yearly (Carapetis et al., 2005). GAS are commonly classified on the basis of the antigenically variable surface M protein, an important virulence factor encoded by the emm gene, of which >200 genotypes have been characterized (McMillan et al., 2013). The last several decades have witnessed an increase in severe invasive forms of GAS infection such as necrotizing fasciitis and streptococcal toxic shock syndrome, largely attributable to the rise of a single globally disseminated hyperinvasive M1T1 (emm1) clone (Aziz and Kotb, 2008; Walker et al., 2014). The ability of M1T1 GAS to produce serious infections defines an intrinsic resistance to the innate immune defenses that normally restrict GAS invasiveness. Indeed, compared to other serotype strains, M1T1 serotype GAS are highly resistant to the cathelicidin-derived peptide LL-37 and to NETs (Lauth et al., 2009), in which cathelicidin is highly abundant. Here we sought to examine the molecular mechanism underlying the cathelicidin resistance of M1T1 GAS and the extent to which it contributes to the increased virulence of these isolates.
RESULTS
GAS is Protected by M1 Protein Binding of LL-37
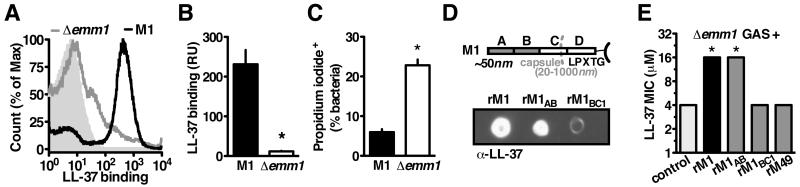
M1T1 GAS lacking M1 protein are more susceptible to killing by human cathelicidin LL-37 (Lauth et al., 2009). M protein is the most abundant protein on the GAS surface (Severin et al., 2007), extending from the bacterial cell surface in the form of hair-like fimbriae (Phillips et al., 1981). One important category of streptococcal antimicrobial peptide resistance mechanisms involves electrostatic repulsion of the cationic peptides away from the bacterial surface (LaRock and Nizet, 2015). However, we found by FACS that M1T1 GAS bound significantly more LL-37 than isogenic Δemm1 mutant bacteria (Figure 1A and 1B). Despite binding more cathelicidin, fewer of the M1-expressing GAS were killed as quantified by propidium iodide uptake (Figure 1C). These results indicate that M1 does not repel LL-37, yet still protects GAS from killing by the antimicrobial peptide.
Figure 1. GAS is Protected by M1 Protein Binding of LL-37.
(A-C) Mid-log wild-type or Δemm1 M1T1 GAS were incubated with 2 μM LL-37 and propidium iodide. (A, B) FACS detection of LL-37 on the bacterial surface with anti-LL-37 antibody indicates Δemm1 GAS binds significantly less LL-37 than wild-type GAS. (C) FACS quantification of cells staining positive for propidium iodide shows significantly more Δemm1 GAS have disrupted cell walls compared to wild-type. (D) Dot blot of pulldown analysis with His-tagged recombinant M protein and LL-37, detected by Western blot with anti-LL-37 antibody. (E) MIC assay of LL-37-susceptible Δemm1 GAS with recombinant M protein to increase resistance. Means ± s.d. (n=3) are shown; *p< 0.05. Figures are representative of at least 3 independent experiments.
We hypothesized that M1 protein could provide an alternative binding site to entrap LL-37 away from the bacterial membrane, thereby preserving bacterial cell integrity. Through pulldown analysis, we found recombinant M1 protein directly bound LL-37 (Figure 1D). Further mapping of this interaction using M1-derived fragments revealed that the region most distal to the bacterial surface, carrying the A region and B repeats (M1AB), was sufficient for binding (Figure 1D). The addition of recombinant M1 or M1AB to cultures of Δemm1 GAS reversed the LL-37 susceptibility phenotype of the mutant (Figure 1E), whereas no protective effect was seen with recombinant M protein from less invasive M49 serotype GAS (Figure 1E). This suggests the linkage of M1 protein to LL-37 resistance is independent of cell alterations due to M1 deletion, but rather derives from direct sequestration of LL-37 by M1 protein.
M1 Binds Immature Cathelicidin to Block Antimicrobial Peptide Generation
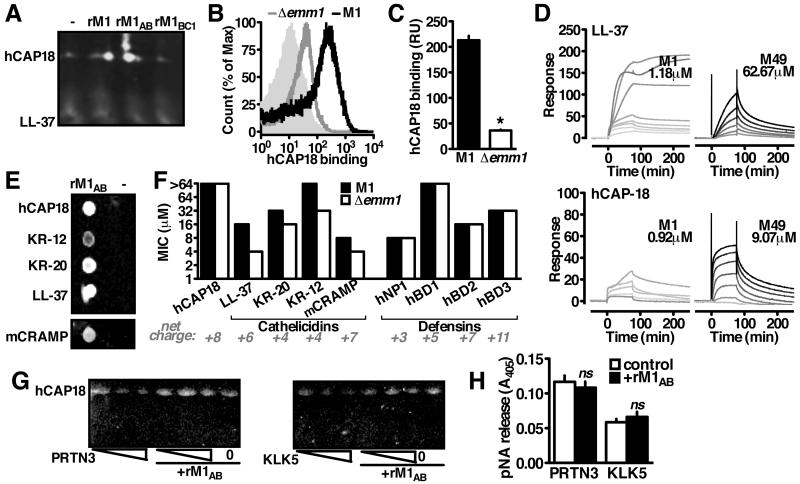
To examine whether M1 sequestration of LL-37 is functional in an infection context, we performed pulldown analysis using whole lysates from neutrophils, the major producers of the human defense peptide (Sørensen et al., 2001). Surprisingly, the major cathelicidin form detected was uncleaved hCAP18 (Figure 2A). This was confirmed by FACS analysis, where recombinant hCAP18 bound the GAS surface in an M1-dependent manner (Figure 2B, 2C). Surface plasmon resonance confirmed a high differential affinity of M1 protein to both LL-37 and hCAP-18. Compared to M49 protein, calculated Kd values for M1 indicated 53-fold stronger binding to LL-37 (1.18 μM vs. 62.67 μM) and 10-fold stronger binding to hCAP-18 (0.92 μM vs. 9.07 μM) (Figure 2D)
Figure 2. M1 Binds Immature Cathelicidin to Block Antimicrobial Peptide Generation.
(A) Interaction between rM1 and cathelicidin from neutrophil lysate by pulldown analysis and Western blot using anti-LL-37 antibody. (B, C) FACS detection of recombinant hCAP18 on the bacterial surface with anti-LL-37 antibody indicates Δemm1 GAS binds significantly less hCAP18 than wild-type GAS. (D) Representative sensorgrams from surface plasmon resonance analysis of M1 or M49 protein binding to immobilized LL-37 or hCAP18. Greyscale indicates concentration of M protein analyte in two-fold dilutions starting from 80 M (black) to 78.125 nM (lightest grey), starting from 5 μM (M1-LL-37), 80 μM (M49-LL-37), 1.25 μM (M1-hCAP18), or (D) 80 μM (M49-hCAP18). (E) Pulldown analysis with rM1 as bait for full-length (hCAP18) or truncated (LL-37, KR-20, KR-12, mCRAMP) forms as shown by dot blot probed with anti-LL-37 antibody (B). (F) Resistance of wild-type and Δemm1 GAS to full-length cathelicidin and the antimicrobial peptides from the cathelicidin (LL-37, KR-20, KR-12, mCRAMP) and defensin (hNP1, hBD1, hBD2, hBD3) families. (G) Cleavage of recombinant hCAP18 by increasing amounts of proteinase-3 (PRTN3) or kalikrein-5 (KLK5) (1, 2, 5 enzymatic units) in the presence or absence of rM1, as visualized by Western blot with anti-LL-37 antibody. (H) Enzymatic activity of proteinase-3 or kalikrein-5 in the presence of rM1. Figures are representative of at least 3 independent experiments.
In addition to binding LL-37 and hCAP18, M1 also bound the peptides KR-20 and KR-12 (Figure 2E), the most active processed forms of cathelicidin expressed by keratinocytes in human skin (Yamasaki et al., 2006). This activity was not limited to human cathelicidin, as the active murine form, mCRAMP, was also bound (Figure 2E). Consistent with binding cathelicidin family peptides, M1 protected GAS from killing by KR-20, KR-12, LL-37, and mCRAMP, but not from the similarly charged defensin-family peptides α-defensin-1 (hNP1), β-defensin-1 (hBD1), β-defensin-2 (hBD2), and β-defensin-3 (hBD3) (Figure 2F).
Unprocessed hCAP18 lacked antimicrobial activity (Figure 2F), consistent with previous reports that only the cleavage products are active (Zaiou et al., 2003). Antimicrobial binding and killing are coupled properties of its cationic charge (Lai and Gallo, 2009), so it was surprising immature cathelicidin interacted with the bacterium without toxicity. M1 protein binding of immature cathelicidin suggested that possibility that the proteolytic maturation of cathelicidin into antimicrobial peptides could be altered. Indeed, M1 inhibited hCAP18 processing by neutrophil proteinase-3 (PRTN3) (Sørensen et al., 2001) and keratinocyte kallikrein-5 (KLK5) (Yamasaki et al., 2006) (Figure 2G). M1 did not inhibit proteinase-3 or kallikrein-5 processing of another substrate (Figure 2H), indicating that M1 is not acting broadly as protease inhibitor. Instead, this suggests that hCAP18 is protected from proteolytic activation when complexed with M1, allowing GAS to inhibit the generation of antimicrobial peptides.
Multipronged Innate Immune Cell Subversion by M1 Through Cathelicidin Sequestration
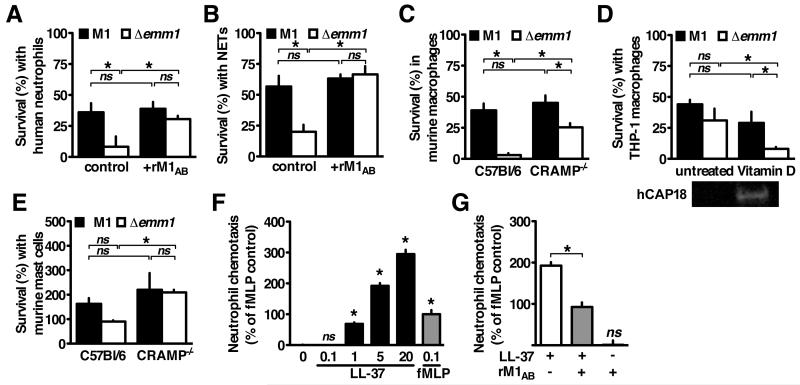
M1 protein is reported to promote GAS survival during interactions with neutrophils (Lauth et al., 2009) and macrophages (Hertzén et al., 2009), two of the major producers of cathelicidin (LaRock and Nizet, 2015). We found that WT GAS was more resistant to human neutrophil killing than the Δemm1 mutant (Figure 3A) and survived better within neutrophil extracellular traps (NETs) (Figure 3B). However, the attenuated survival of the Δemm1 mutant incubated with neutrophils or NETs was reversed by the addition of exogenous recombinant M1 protein (Figure 3A, 3B). In neither instance did addition of rM1 promote further survival of WT GAS, indicating that the native expression of M1 is sufficient to afford complete protection against neutrophil-expressed cathelicidin. These findings indicate that M1 does not have to be expressed on the bacterial surface to be protective, consistent with the molecular mechanism of cathelicidin sequestration.
Figure 3. Multipronged Innate Immune Cell Subversion by M1 Through Cathelicidin Sequestration.
(A-B) Human neutrophils were infected with GAS for 2 h, (B) after 4 h pretreatment with 25nM PMA to induce NET formation. (C-E) Cells derived from C57Bl/6 or CRAMP−/− mice or (E) culture were primed with 100 ng/mL LPS (C, D) or 20 nM vitamin D (active 1,25D3 form) (E) for 18 h then infected with wild-type M1 or Δemm1 GAS for 2 h. (F-G) Human neutrophils were seeded in the upper chamber of a 3 m transwell with LL-37, fMLP, and/or recombinant M1 protein in the lower chamber, and migration measured by myeloperoxidase activity in the lower chamber after 1 h. Means ± s.d. (n=3) are shown; *p< 0.05. Figures are representative of at least 3 independent experiments.
To determine if M1 inhibition of cathelicidin contributes to GAS resistance to macrophage killing, we infected macrophages derived from wild-type (C57Bl/6) and cathelicidin-deficient (CRAMP−/−) mice. Compared to the WT GAS, the Δemm1 mutant had markedly attenuated survival in wild-type macrophages, but similar growth in CRAMP−/− macrophages (Figure 3C). In human cells, vitamin D strongly induces cathelicidin expression (Liu et al., 2006). We found that M1 mutant GAS became hyper-susceptible to killing by human THP-1 macrophages when cathelicidin expression was amplified by vitamin D treatment (Figure 3D). Cathelicidin is also abundantly produced by mast cells and contributed to killing of the cathelicidin-susceptible serotype M49 GAS in the mouse model (Di Nardo et al., 2008). We found that M1 promoted GAS resistance to killing by murine mast cell cells in a cathelicidin-dependent manner (Figure 3E).
In addition to microbial killing, cathelicidin cleavage products further amplify the innate immune response by stimulating chemotaxis of neutrophils to the foci of infection (Yang et al., 2000). We tested the effect of M1 protein on LL-37-mediated chemotaxis using a transwell system. LL-37 and the positive control N-formyl-Met-Leu-Phe (fMLP) both stimulated robust chemotaxis of neutrophils (Figure 3F). rM1 did not stimulate or inhibit chemotaxis on its own, but did inhibit LL-37-mediated migration (Figure 3G). Chemotaxis inhibition by the M1 protein further indicates interfering with cathelicidin function is a virulence strategy that can promote bacterial survival by several modalities.
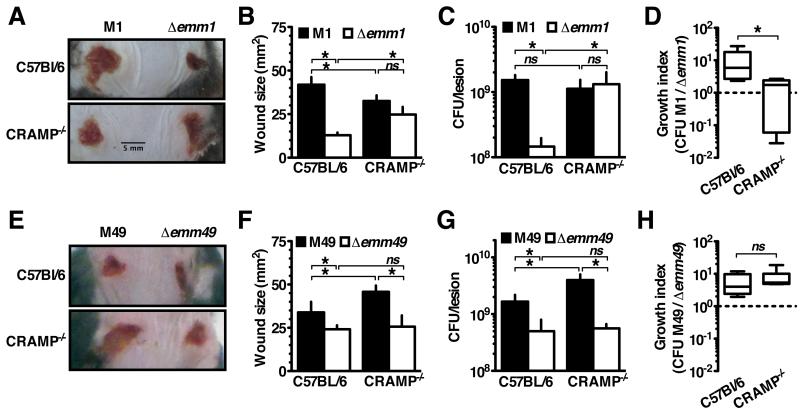
GAS Inhibition of Cathelicidin Permits Invasive Infection
Our in vitro analyses suggest that M1 protein binding of cathelicidin could promote GAS innate immune resistance in several manners. To examine the contribution of cathelicidin sequestration by the M1 protein on GAS in vivo, we used a subcutaneous wound infection model with C57Bl/6 and CRAMP−/− littermate mice and followed the development of necrotic skin ulcers at the site of GAS inoculation. In WT mice, WT M1 GAS produced larger lesions than the Δemm1 mutant, consistent with the known virulence role of the M1 protein (Figure 4A, 4B). Remarkably, in CRAMP-deficient (−/−) mice Δemm1 GAS was able to form lesions of size comparable to WT GAS (Figure 4A, 4B). This reversal of attenuation in CRAMP−/− mice was not seen for Δemm49 GAS (Figure 4E, 4F), which agrees with the observation that M1 but not M49 binds and inhibits CRAMP. Interestingly, M49 GAS formed larger lesions in CRAMP−/− mice (Figure 4E, 4F), whereas M1 GAS produced similar sized lesions in WT and cathelicidin-deficient animals (Figure 4A, 4B), suggesting the M1 protein effectively counteracts the cathelicidin contribution to innate defense in this localized GAS infection model.
Figure 4. GAS Inhibition of Cathelicidin Permits Invasive Infection.
(A-H) Mice were infected subcutaneously with the GAS strains indicated for 72 h. (A, E) GAS mutant for M protein (Δemm1; Δemm49) formed smaller lesions in cathelicidin-expressing mice (wild-type, C57Bl/6), but the capacity to form lesions was restored for Δemm1 in CRAMP−/− mice. Average lesion sizes enumerated in (B, F) demonstrate these differences are significant. (C, G), Lesions were excised, homogenized, and CFU enumerated by dilution plating. (D, H) Growth index, measuring growth of wild-type bacteria compared to emm-mutants, showed wild-type GAS outcompeting mutant bacteria 10-fold except for in CRAMP−/− mice where M1 and M1 Δemm1 grew comparably. Means ± s.d. (n=5) are shown; *p< 0.05. Figures are representative of at least 3 independent experiments.
When bacteria were harvested for colony forming unit (CFU) enumeration 72 h after infection, the Δemm1 GAS recovered from WT mice were comparable to the initial inoculum, whereas WT M1 GAS had proliferated over an order of magnitude (Figure 4C). Consistent with the lesion size data, in vivo proliferation of Δemm1 GAS was restored to WT levels in CRAMP−/− mice (Figure 4C). Together, these data suggest that while M proteins have many virulence functions, cathelicidin inhibition is the major virulence contribution of the M1 protein in this model. Although Δemm49 was similarly attenuated in WT mice, its growth was not similarly restored in CRAMP−/− mice (Figure 4G). This indicates the attenuation reversal of Δemm1 is specifically due to the ability of M1 to inhibit cathelicidin, and not a more generalized immune defect of CRAMP−/− mice. Highlighting these differences in another way, the growth index of WT M1 GAS was greater than the isogenic Δemm1 mutant only in WT mice where cathelicidin is expressed (Figure 4D), whereas M49 contribution to virulence was independent of host cathelicidin expression (Figure 4H).
DISCUSSION
Antimicrobial peptides provide a countermeasure against bacteria that is conserved across all kingdoms of life. Mice lacking cathelicidin antimicrobial peptides are more susceptible to infection (Nizet et al., 2001), and qualitative defects in cathelicidin expression or activity play a role in the increased infection rates of humans with cystic fibrosis (Goldman et al., 1997), morbus Kostmann (Pütsep et al., 2002), and chronic vitamin D deficiency (Liu et al., 2006). Conversely, dysregulated and excessive levels of cathelicidin expression can be damaging to the host and a driver of diseases such as rosacea (Yamasaki et al., 2007), hidradenitis suppurativa (Emelianov et al., 2012), and cigarette smoke-associated chronic obstructive pulmonary disease (Sun et al., 2014). Cathelicidin expression may be limited because of these proinflammatory liabilities to levels for which resistance by pathogens is possible.
Our results show that one pathogenic bacterial clone, hypervirulent M1T1 GAS, has developed high-level cathelicidin resistance. Indeed, M1 GAS produced equivalent lesions and bacterial loads in WT and CRAMP−/− mice, indicating it is fully resistant to cathelicidin in vivo which has provided it a significant virulence benefit. In contrast, M49 GAS has not evolved high-level resistance to cathelicidin. It causes more severe disease in CRAMP−/− mice than WT, as is true for other pathogens tested (Bergman et al., 2006; Chromek et al., 2006; Huang et al., 2007; Kovach et al., 2012; Nizet et al., 2001). Therefore physiological levels of cathelicidin are restrictive for other GAS, but M1 protein allows the resistance of hypervirulent M1T1 GAS to levels above those achieved in vivo.
M1 protein provides a multifaceted cathelicidin resistance advantage to GAS. Specifically, M1 directly binds LL-37 and other cathelicidin derivatives. This sequestration is sufficient to double or quadruple the concentration of these peptides required to kill the bacterium. Resistance to these peptides carries over to the interaction of GAS with cathelicidin-expressing immune cells such as macrophages, mast cells, and neutrophils. Moreover, M1 protein also inhibits cathelicidin-induced chemotaxis, further promoting GAS evasion of the immune response.
Interestingly, M1 protein bound the uncleaved storage form of cathelicidin in addition to the mature antimicrobial peptides. However, because uncleaved cathelicidin and its activating proteases can be found extracellularly (Sørensen et al., 1999), this may represent another facet of the antimicrobial evasion strategy of GAS. Cathelicidin bound by M1 protein was no longer cleaved by activating proteases, maintaining it in a non-antimicrobial form. By acting upstream of the activation of cathelicidin, GAS is able to pre-empt bacterial killing. In contrast to this two-pronged mechanism we describe for M1 protein, other pathogens are only known to resist antimicrobial peptides through sequestration, degradation, and masking or altering their cell surface (LaRock and Nizet, 2015). Identification of additional antimicrobial peptide-binding virulence factors may reveal this mechanism to be prevalent, with the microbe agnostic to the cleavage state of cathelicidin. Understanding the molecular mechanisms by which microbial pathogens control cathelicidin may provide targets to sensitize the pathogen to innate immune clearance, or conversely, strategies to co-opt the same cathelicidin inhibitors for use in immune diseases such as rosacea where cathelicidin overexpression is implicated.
EXPERIMENTAL PROCEDURES
Bacterial Culture
GAS M1T1 5448, M49 NZ131, and their isogenic Δemm mutants have been previously described (Lauth et al., 2009). GAS strains were routinely propagated at 37°C on Todd-Hewitt media (THB, Difco). GAS grown overnight at 37°C were diluted 1:40 into Todd-Hewitt media and grown statically at 37°C to OD600 0.400. Bacteria washed in PBS and diluted to a multiplicity of infection (MOI) of 10 for in vitro experiments. For minimum inhibitory concentration (MIC) assays, 2×105 bacteria were diluted into serum-free DMEM containing phenol red and 10% THB and the minimum antimicrobial concentration that prevented growth of bacteria after a 24 h incubation was noted.
Protein protocols
Expression and purification of rM1, rM1AB, rM1BC1, and rM49 has been described previously (Lauth et al., 2009; Macheboeuf et al., 2011; McNamara et al., 2008). The cathelicidin peptides LL-37, KR-20, KR-12, and mCRAMP were made by de novo synthesis (Anaspec). The full coding sequence of human cathelicidin hCAP18 was chemically synthesized (GenScript) and inserted into the expression vector pET28a. The protein was expressed using standard protocols, and native purified by standard protocols in buffer made of 10 mM imidazole, 0.3 M NaCl, 20mM Na2HPO4 pH 7.4 using cobalt chelate resin (Thermo Fisher). Pulldowns were performed by fixing 5 μg recombinant 6×His tagged M1 protein onto cobalt chelate resin as bait (Thermo Fisher), incubating with 5 μg cathelicidin, and eluting with 100 mM imidazole, with three washes between each step, with the same buffer at all steps (5 mM imidazole, 100 mM NaCl, 20 mM Tris pH 7). Samples were run by SDS-PAGE or dot-blotted directly onto PVDF and immunobloted for LL-37 or CRAMP (Santa Cruz) with IR-conjugated secondary antibodies (LI-COR), and visualized on an Odyssey infrared scanner (LI-COR). Proteolysis assays were carried out with 1 μg cathelicidin and M protein, with the indicated amount of Proteinase-3 or Kallikrein-5 (VWR) and visualized by western blot, or VLD-pNA or AAPV-pNA (Enzo) was added and hydrolysis monitored by an increase in OD405 (Spectramax).
Binding kinetics were analyzed by surface plasmon resonance using a Biacore T100 (GE Healthcare). A C1 sensor chip was activated with 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide and N-hydroxysuccinimide. LL-37 or CAP18 in 10 mM sodium acetate pH 5.5 were injected at 10 μL/min for 420 s. Remaining reactive groups were quenched using 1 M ethanolamine. In parallel, a reference flow cell was activated and quenched without ligand immobilization. M proteins in PBS, 0.005% Tween-20 were injected at 30 μL/min for 75 s. After at least 180 s of dissociation the chip was regenerated using 10 mM glycine-HCl pH 1.7. Reference and buffer only values were subtracted and kinetic analysis was performed using Biacore Evalution Software (GE Healthcare). Kd values were calculated using a steady-state affinity model. Each experiment was performed twice with similar results.
Cell Culture
Cell lines were cultured in RPMI (Gibco) containing 10% fetal bovine serum (Gibco) at 37°C in 5% CO2 and seeded at 2×105 cells/mL. THP-1 cells were originally obtained from ATCC and treated with phorbol 12-myristate 13-acetate (Sigma) 72 h pre-infection. Murine macrophages, mast cells, and neutrophils were differentiated by standard protocols from the femur exudates of wild-type C57Bl/6 or isogenic CRAMP−/− mice (Nizet et al., 2001). Human neutrophils were isolated from healthy donors using PolyMorphPrep (Axis-Shield) from healthy donors in accordance with the UC San Diego Human Research Protections Program. For total killing, the cells were lysed with 0.05% Triton-X100 for 10 min before dilution plating for CFU. In intracellular survival experiments, the media was replaced with RPMI containing 100 μg/mL gentamicin 30 min before lysis and plating. For NET assays neutrophils were stimulated with 25nM phorbol 12-myristate 13-acetate (Sigma) 4 h pre-infection. For chemotaxis assays, 1×106 cells were seeded in the upper chamber of a 3-μm pore Transwell inserts (Corning), with the indicated concentrations of fMLP (Sigma), LL-37 (Anaspec), and rM1 in RPMI in the lower chamber. Following incubation at for 1 h at 37°C with 5% CO2, transwell inserts were removed and cells migrated to the lower chamber were lysed via addition of 0.1% Triton-X 100. To quantify chemotaxis, the chromogenic elastase substrate N-methoxysuccinyl-Ala-Ala-Pro-Val p-nitroanilide was added to lysate samples (1 mM final); following a 30 min incubation at room temperature, absorbance at 405 nm was measured using a SpectraMax plate reader (Molecular Devices).
Cell imaging
For flow cytometery, samples were prepared by growing GAS by the standard protocol followed by 30 min incubation with 2 μM LL-37. Cells were washed, blocked 30 min with 10% BSA (Sigma), probed with anti-LL-37 (Santa Cruz) and Alexa-dye conjugated secondary antibody (Invitrogen), and run on a FACSCalibur system (BD). Bacterial cells were gated according to forward and side scatter and the fluorescence intensity measured for a total of 50,000 bacteria. Negative controls of bacteria untreated by LL-37, primary antibody, or secondary antibody were included and data was analyzed with FlowJo (Tree Star). Cell permeability was monitored in parallel by staining by 10 mg/ml propidium iodide. Immunofluorescence images were collected using an AxioObserver D1 microscope equipped with an LD A-Plan 20×/0.35 Ph1 objective (Zeiss) microscope and processed in ImageJ.
Animal Experiments
The UCSD Institutional Animal Care and Use Committee approved all use and procedures. Eight- to ten-week old C57BL/6 and littermate CRAMP−/− mice were infected subcutaneously with 1×108 (M1) or 6×108 (M49) CFU in 100 μL of PBS. Lesions were imaged daily and surface area quantified using ImageJ software. At 72 h lesions were excised, homogenized, and dilution plated onto Todd-Hewitt agar plates for enumeration of bacterial CFU.
Statistical Analysis
Statistical significance (*; p<0.05) was calculated by unpaired Student’s t-test (GraphPad Prism), unless otherwise indicated. Data are representative of at least three independent experiments.
Supplementary Material
HIGHLIGHTS.
Hypervirulent M1 group A Streptococcus (GAS) resists innate immune clearance
The surface-expressed GAS M1 protein sequesters human cathelicidin LL-37
M1 protein sequestration of immature cathelicidin blocks its activation
The pathogenesis of GAS necrotizing fasciitis requires cathelicidin inhibition
ACKNOWLEDGMENTS
We would like to thank the members of the Ghosh and Nizet labs for their valuable input and Felix Bröcker for assistance with kinetic analysis. This work was supported in part by a fellowship from the A.P. Giannini Foundation (C.N.L.), the German Academic Exchange Service (DAAD) (S.D.), and National Institute of Health grants AI052453 (R.L.G. and V.N), AI096837 (P.G. and V.N), and AI077780 (V.N.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
C.N.L., P.G., and V.N. formulated the original hypothesis, designed the study, and analyzed results. C.N.L., S.D., J.T., T.J., R.C., and J.O. performed experiments. B.L. and R.L.G. provided novel reagents. C.N.L. and V.N. wrote the manuscript, and all authors reviewed the manuscript, data, and conclusions before submission.
We declare no conflicts in interest.
REFERENCES
- Alalwani SM, Sierigk J, Herr C, Pinkenburg O, Gallo R, Vogelmeier C, Bals R. The antimicrobial peptide LL-37 modulates the inflammatory and host defense response of human neutrophils. Eur J Immunol. 2010;40:1118–1126. doi: 10.1002/eji.200939275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Kotb M. Rise and persistence of global M1T1 clone of Streptococcus pyogenes. Emerg Infect Dis. 2008;14:1511–1517. doi: 10.3201/eid1410.071660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman P, Johansson L, Wan H, Jones A, Gallo RL, Gudmundsson GH, Hökfelt T, Jonsson A-B, Agerberth B. Induction of the antimicrobial peptide CRAMP in the blood-brain barrier and meninges after meningococcal infection. Infect Immun. 2006;74:6982–6991. doi: 10.1128/IAI.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Chromek M, Slamova Z, Bergman P, Kovacs L, Podracka L.u., Ehren I, Hokfelt T, Gudmundsson GH, Gallo RL, Agerberth B, et al. The antimicrobial peptide cathelicidin protects the urinary tract against invasive bacterial infection. Nat Med. 2006;12:636–641. doi: 10.1038/nm1407. [DOI] [PubMed] [Google Scholar]
- Di Nardo A, Yamasaki K, Dorschner RA, Lai Y, Gallo RL. Mast cell cathelicidin antimicrobial peptide prevents invasive group A Streptococcus infection of the skin. J Immunol. 2008;180:7565–7573. doi: 10.4049/jimmunol.180.11.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorschner RA, Pestonjamasp VK, Tamakuwala S, Ohtake T, Rudisill J, Nizet V, Agerberth B, Gudmundsson GH, Gallo RL. Cutaneous injury induces the release of cathelicidin anti-microbial peptides active against group A Streptococcus. J Invest Derm. 2001;117:91–97. doi: 10.1046/j.1523-1747.2001.01340.x. [DOI] [PubMed] [Google Scholar]
- Emelianov VU, Bechara FG, Gläser R, Langan EA, Taungjaruwinai WM, Schröder JM, Meyer KC, Paus R. Immunohistological pointers to a possible role for excessive cathelicidin (LL-37) expression by apocrine sweat glands in the pathogenesis of hidradenitis suppurativa/acne inversa. Brit J Derm. 2012;166:1023–1034. doi: 10.1111/j.1365-2133.2011.10765.x. [DOI] [PubMed] [Google Scholar]
- Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human β-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–560. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- Hertzén E, Johansson L, Wallin R, Schmidt H, Kroll M, Rehn AP, Kotb M, Mörgelin M, Norrby-Teglund A. M1 protein-dependent intracellular trafficking promotes persistence and replication of Streptococcus pyogenes in macrophages. J Innate Immun. 2009;2:534–545. doi: 10.1159/000317635. [DOI] [PubMed] [Google Scholar]
- Ho Wong J, Juan Ye X, Bun Ng T. Cathelicidins: peptides with antimicrobial, immunomodulatory, anti-inflammatory, angiogenic, anticancer and procancer activities. Curr Prot Peptide Sci. 2013;14:504–514. doi: 10.2174/13892037113149990067. [DOI] [PubMed] [Google Scholar]
- Huang LC, Reins RY, Gallo RL, McDermott AM. Cathelicidin-deficient (Cnlp−/−) mice show increased susceptibility to Pseudomonas aeruginosa keratitis. Invest Ophthalmol Visual Sci. 2007;48:4498–4508. doi: 10.1167/iovs.07-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach MA, Ballinger MN, Newstead MW, Zeng X, Bhan U, Yu F.-s., Moore BB, Gallo RL, Standiford TJ. Cathelicidin-related antimicrobial peptide is required for effective lung mucosal immunity in gram-negative bacterial pneumonia. J Immunol. 2012;189:304–311. doi: 10.4049/jimmunol.1103196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–141. doi: 10.1016/j.it.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock CN, Nizet V. Cationic antimicrobial peptide resistance mechanisms of streptococcal pathogens. Biochem Biophys Acta. 2015 Feb 17; doi: 10.1016/j.bbamem.2015.02.010. pii: S0005-2736(15)00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauth X, von Köckritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Inate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PT, Stenger S, Li H, Wenzel L, Tan BH, Krutzik SR, Ochoa MT, Schauber J, Wu K, Meinken C, et al. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science. 2006;311:1770–1773. doi: 10.1126/science.1123933. [DOI] [PubMed] [Google Scholar]
- Macheboeuf P, Buffalo C, Fu C.-y., Zinkernagel AS, Cole JN, Johnson JE, Nizet V, Ghosh P. Streptococcal M1 protein constructs a pathological host fibrinogen network. Nature. 2011;472:64–68. doi: 10.1038/nature09967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DJ, Drèze PA, Vu T, Bessen DE, Guglielmini J, Steer AC, Carapetis JR, Van Melderen L, Sriprakash KS, Smeesters PR. Updated model of group A Streptococcus M proteins based on a comprehensive worldwide study. Clin Microbiol Infect. 2013;19:E222–E229. doi: 10.1111/1469-0691.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara C, Zinkernagel AS, Macheboeuf P, Cunningham MW, Nizet V, Ghosh P. Coiled-coil irregularities and instabilities in group A Streptococcus M1 are required for virulence. Science. 2008;319:1405–1408. doi: 10.1126/science.1154470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Lopez-Garcia B, Braff M, Dorschner RA, Gallo RL. Postsecretory processing generates multiple cathelicidins for enhanced topical antimicrobial defense. J Immunol. 2004;172:3070–3077. doi: 10.4049/jimmunol.172.5.3070. [DOI] [PubMed] [Google Scholar]
- Neumann A, Berends E, Nerlich A, Molhoek EM, Gallo RL, Meerloo T, Nizet V, Naim HY, von Kockritz-Blickwede M. The antimicrobial peptide LL-37 facilitates the formation of neutrophil extracellular traps. Biochemical J. 2014;464:3–11. doi: 10.1042/BJ20140778. [DOI] [PubMed] [Google Scholar]
- Nizet V, Ohtake T, Lauth X, Trowbridge J, Rudisill J, Dorschner RA, Pestonjamasp V, Piraino J, Huttner K, Gallo RL. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature. 2001;414:454–457. doi: 10.1038/35106587. [DOI] [PubMed] [Google Scholar]
- Phillips GN, Flicker PF, Cohen C, Manjula BN, Fischetti VA. Streptococcal M protein: alpha-helical coiled-coil structure and arrangement on the cell surface. Proc Natl Acad Sci USA. 1981;78:4689–4693. doi: 10.1073/pnas.78.8.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pütsep K, Carlsson G, Boman HG, Andersson M. Deficiency of antibacterial peptides in patients with morbus Kostmann: an observation study. Lancet. 2002;360:1144–1149. doi: 10.1016/S0140-6736(02)11201-3. [DOI] [PubMed] [Google Scholar]
- Severin A, Nickbarg E, Wooters J, Quazi SA, Matsuka YV, Murphy E, Moutsatsos IK, Zagursky RJ, Olmsted SB. Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J Bacteriol. 2007;189:1514–1522. doi: 10.1128/JB.01132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen O, Bratt T, Johnsen AH, Madsen MT, Borregaard N. The human antibacterial cathelicidin, hCAP-18, is bound to lipoproteins in plasma. J Biol Chem. 1999;274:22445–22451. doi: 10.1074/jbc.274.32.22445. [DOI] [PubMed] [Google Scholar]
- Sørensen OE, Follin P, Johnsen AH, Calafat J, Tjabringa GS, Hiemstra PS, Borregaard N. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood. 2001;97:3951–3959. doi: 10.1182/blood.v97.12.3951. [DOI] [PubMed] [Google Scholar]
- Sun C, Zhu M, Yang Z, Pan X, Zhang Y, Wang Q, Xiao W. LL-37 secreted by epithelium promotes fibroblast collagen production: a potential mechanism of small airway remodeling in chronic obstructive pulmonary disease. Lab Invest. 2014;94:991–1002. doi: 10.1038/labinvest.2014.86. [DOI] [PubMed] [Google Scholar]
- Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev. 2014;27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Di Nardo A, Bardan A, Murakami M, Ohtake T, Coda A, Dorschner RA, Bonnart C, Descargues P, Hovnanian A. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat Med. 2007;13:975–980. doi: 10.1038/nm1616. [DOI] [PubMed] [Google Scholar]
- Yamasaki K, Schauber J, Coda A, Lin H, Dorschner RA, Schechter NM, Bonnart C, Descargues P, Hovnanian A, Gallo RL. Kallikrein-mediated proteolysis regulates the antimicrobial effects of cathelicidins in skin. FASEB J. 2006;20:2068–2080. doi: 10.1096/fj.06-6075com. r. [DOI] [PubMed] [Google Scholar]
- Yang D, Chen Q, Schmidt AP, Anderson GM, Wang JM, Wooters J, Oppenheim JJ, Chertov O. LL-37, the neutrophil granule–and epithelial cell–derived cathelicidin, utilizes formyl peptide receptor-like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192:1069–1074. doi: 10.1084/jem.192.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiou M, Nizet V, Gallo RL. Antimicrobial and protease inhibitory functions of the human cathelicidin (hCAP18/LL-37) prosequence. J Invest Derm. 2003;120:810–816. doi: 10.1046/j.1523-1747.2003.12132.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.