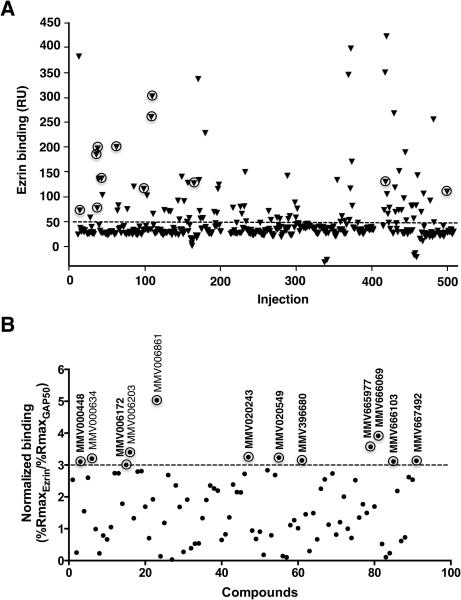
Figure 1.
Screening of MMV400 library by surface plasmon resonance technology for direct binding to ezrin. (A) Purified recombinant ezrin protein was immobilized on a sensor chip and small molecules were individually injected over the surface at a single concentration in a Biacore 3000 instrument. A negative control protein GAP50 was also immobilized on a neighboring flow cell in the same sensor chip to eliminate false positives resulting from non-specific protein binding. The data points represented with triangles encircled with rings represent the primary hits with relatively high specific binding for ezrin identified through filtering of the data as explained in panel B. (B) Compound filtering was implemented as a mean to prioritize the hits based on their specific binding to ezrin over GAP50, which yielded a total of 12 primary hits. The analyte binding capacity or theoretical maximum response (Rmax for maximum response) of each compound were calculated for both ezrin and GAP50. The relative binding of each compound (response units, RU) to ezrin and GAP50 were normalized to Rmax, which was taken as 100%. Any molecule demonstrating a threshold of 3-fold difference in relative binding to ezrin in terms of normalized response with respect to the negative control protein GAP50 was then selected as a primary hit. 9 compounds out of 12 primary hits used in this study are shown in bold.