Fig 4. Summary of EAST interactions with insulator proteins in the yeast two-hybrid assay.
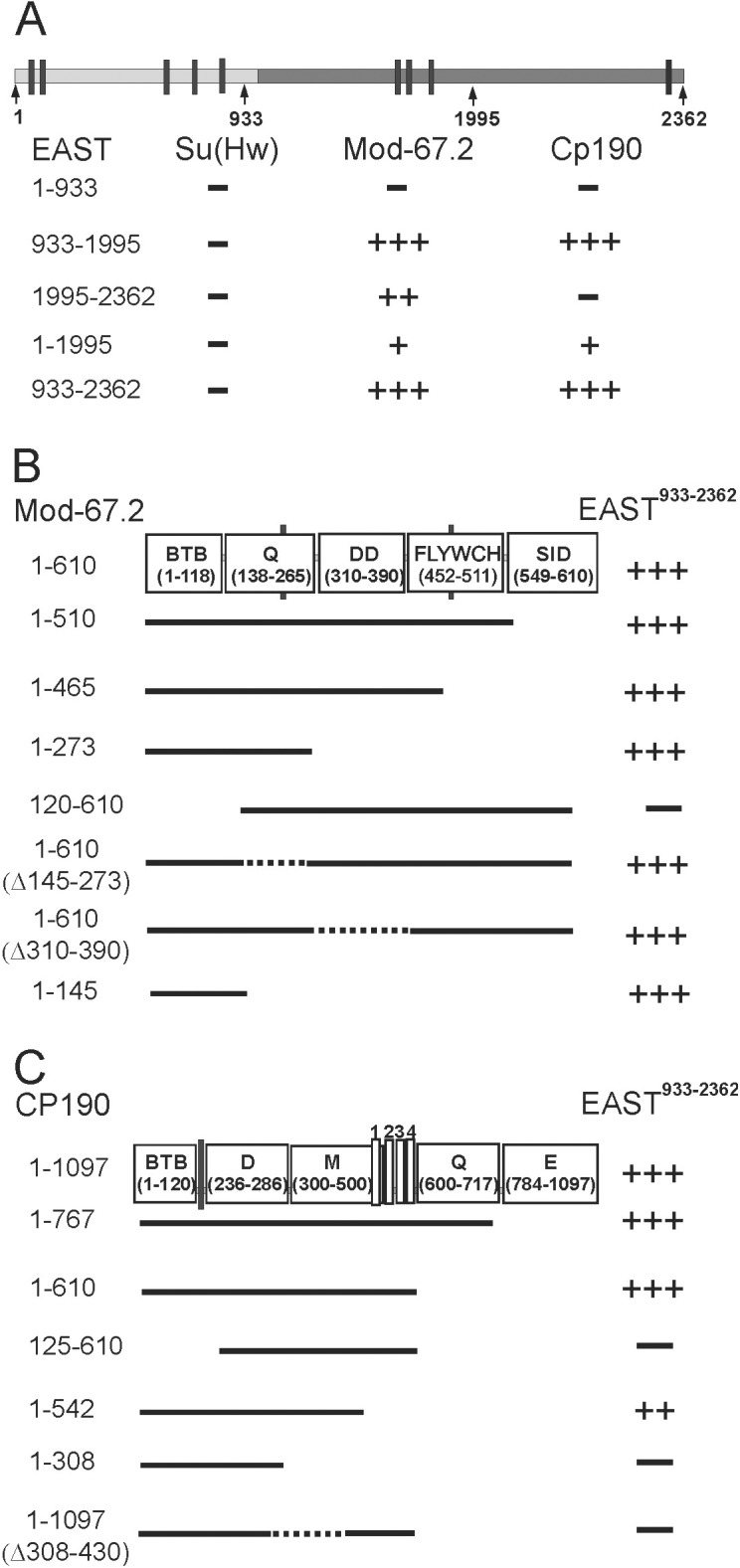
(A) The results of testing parts of EAST for the interaction with the Su(Hw), CP190 and Mod(mdg4)-67.2 proteins. The scheme shows the structure of full-length EAST, with light and dark gray shading indicating its unstructured and highly structured regions, respectively. For identification of the different EAST protein signatures, InterProScan tool from EMBL-EBI resource was used. It was found that EAST possesses highly structured middle and C terminal regions, in contrast to the unstructured N terminal region. (B) The results of testing Mod(mdg4)-67.2 domains for the interaction with EAST. The scheme shows Mod(mdg4)-67.2 domains with the numbers of amino acid residues included in the corresponding protein product (in parentheses). (C) The results of testing CP190 domains for the interaction with EAST. The scheme shows CP190 domains with the numbers of amino acid residues included in the corresponding protein product (in parentheses), four C2H2 zinc fingers (white rectangles), and the nuclear localization signal (black bar). Figures in the schemes and on the left are the numbers of amino acid residues. The plus signs indicate the relative strength of two-hybrid interaction; the minus sign, the absence of interaction (See S6 Fig).
